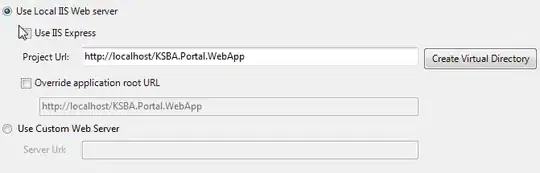
I have the following problem. If you take a look at the image below, you see a microscopic image of a circular strand of DNA, called a teardrop. An initial skeletonization (morphology.skeletonize) yielded the trace in red. The trace is an nd-array of shape (N, 2), where N is the number of points in my trace. In orange, the strand is traced after I applied some sort of improvement algorithm, which I designed myself. Now, as you can see, around the bright region, the trace twists a little bit and doesn't resemble the real shape. To work around that, I want to do the following:
- Define a radius around the blue dot which is fixed (i.e. 5 nanometres)
- Remove all points of the trace that lie within that radius (my problem)
- After those points are removed, connect the blue dot with the first point that lies outside the circle
In the end, I don't know how I can tell Python that for a given array of coordinates, I want to delete all those that lie within an area of my image. That area is, as explained, a circle around the blue dot.