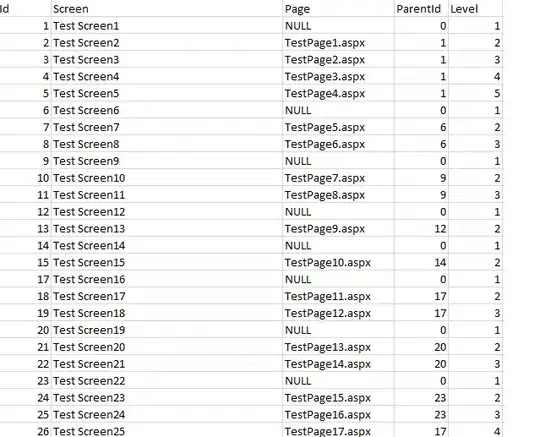
I'm trying to create a RidgePlot of this data:
long_data <- structure(list(Cell_Type = structure(c(1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 6L, 6L,
6L, 6L, 7L, 7L, 7L, 7L, 8L, 8L, 8L, 8L, 9L, 9L, 9L, 9L, 10L,
10L, 10L, 10L), .Label = c("CD4..T.cells", "CD4..Tcm", "CD8..Tcm",
"CD8..Tem", "Class.switched.memory.B.cells", "Monocytes", "MPP",
"naive.B.cells", "NK.cells", "Plasma.cells"), class = "factor"),
Sample = structure(c(1L, 2L, 3L, 4L, 1L, 2L, 3L, 4L, 1L,
2L, 3L, 4L, 1L, 2L, 3L, 4L, 1L, 2L, 3L, 4L, 1L, 2L, 3L, 4L,
1L, 2L, 3L, 4L, 1L, 2L, 3L, 4L, 1L, 2L, 3L, 4L, 1L, 2L, 3L,
4L), .Label = c("Sample1", "Sample2", "Sample3", "Sample4"
), class = "factor"), Percentage = c(39, 35.1, 12.7405962,
4.995628, 11.4, 10.9, 8.9116355, 10.1955, 4.5, 6.1, 18.2643388,
7.005608, 5.9, 6, 4.9746425, 10.288099, 2.8, 2.8, 13.7408777,
19.657708, 18.8, 21.4, 5.7131852, 19.657708, 0.6, 0.8, 0.2308895,
18.295758, 7, 6.2, 7.210278, 3.383666, 9.9, 9.8, 16.9838566,
9.087761, 0.1, 1, 11.2297, 10.511573), outlier = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L,
1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L,
1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L), .Label = c("no", "yes"
), class = "factor")), class = "data.frame", row.names = c(NA,
-40L))
column outlier is a factor of two levels "yes" and "no" but when I create the ggplot it is completely ignoring all the rows with "yes" for outlier.
Here's an example of the ggplot I'm getting. I seem to be missing something here because the expected output is a legend with "no" and "yes" and points with two different colors.
I seem to be missing something here because the expected output is a legend with "no" and "yes" and points with two different colors.
This is the code to produce it:
ggplot(long_data, aes(x=Percentage, y=reorder(Cell_Type,Percentage))) +
geom_density_ridges(scale=2, alpha=0.2)+
geom_density_ridges(alpha=0, color=NA, jittered_points=T, point_alpha=1, aes(point_color=outlier)) +
xlab("Percentage") + ylab("Cell Type")
what am I missing here?