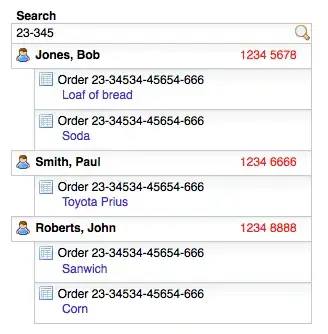
I have been trying to make a plot that shows how often a specific world region (Western Europe, North America, etc...) has been covered in an issue of a certain journal.
I'd like the x-axis to display the issue and the y-axis to display the count of times a region has been covered in that issue. I would like to create a stacked area chart, so I can see which region has been covered the most in which issue, and how these numbers change over time.
My data looks like this:
The observations that include data on other world regions (WE = Western Europe, EE = Eastern Europe, etc.) are not shown in the picture, but follow further down.
My code looks like this:
wreg_by_issue_plot <- ggplot(data = cov_by_issue_long, aes(x = issue, y = count, fill = wreg)) +
geom_area()
According to this tutorial my code should work, but it doesn't. Instead, I get the following outcome:

I know, the x-axis is still messy, but I can take care of that later. What I wonder is, why are the areas not being displayed?