If the sample has repeated values, this implies that the underlying distribution is not continuous. In the data that you show to illustrate the issue, we can see a Dirac distribution on the left. The kernel smoothing might be applied for such data, but with care. Indeed, to approximate such data, we might use a kernel smoothing where the bandwidth associated to the Dirac is zero. However, in most KDE methods, there is only one single bandwidth for all kernel atoms. Moreover, the various rules used to compute the bandwidth are based on some estimation of the rugosity of the second derivative of the PDF of the distribution. This cannot be applied to a discontinuous distribution.
We can, however, try to separate the sample into two sub-samples:
- the sub-sample(s) with replications,
- the sub-sample with unique realizations.
(This idea has already been mentionned by johanc).
Below is an attempt to perform this classification. The np.unique method is used to count the occurences of the replicated realizations. The replicated values are associated with Diracs and the weight in the mixture is estimated from the fraction of these replicated values in the sample. The remaining realizations, uniques, are then used to estimate the continuous distribution with KDE.
The following function will be useful in order to overcome a limitation with the current implementation of the draw method of Mixtures with OpenTURNS.
def DrawMixtureWithDiracs(distribution):
"""Draw a distributions which has Diracs.
https://github.com/openturns/openturns/issues/1489"""
graph = distribution.drawPDF()
graph.setLegends(["Mixture"])
for atom in distribution.getDistributionCollection():
if atom.getName() == "Dirac":
curve = atom.drawPDF()
curve.setLegends(["Dirac"])
graph.add(curve)
return graph
The following script creates a use-case with a Mixture containing a Dirac and a gaussian distributions.
import openturns as ot
import numpy as np
distribution = ot.Mixture([ot.Dirac(-3.0),
ot.Normal()], [0.5, 0.5])
DrawMixtureWithDiracs(distribution)
This is the result.
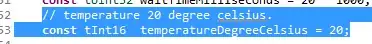
Then we create a sample.
sample = distribution.getSample(100)
This is where your problem begins. We count the number of occurences of each realizations.
array = np.array(sample)
unique, index, count = np.unique(array, axis=0, return_index=True,
return_counts=True)
For all realizations, replicated values are associated with Diracs and unique values are put in a separate list.
sampleSize = sample.getSize()
listOfDiracs = []
listOfWeights = []
uniqueValues = []
for i in range(len(unique)):
if count[i] == 1:
uniqueValues.append(unique[i][0])
else:
atom = ot.Dirac(unique[i])
listOfDiracs.append(atom)
w = count[i] / sampleSize
print("New Dirac =", unique[i], " with weight =", w)
listOfWeights.append(w)
The weight of the continuous atom is the complementary of the sum of the weights of the Diracs. This way, the sum of the weights will be equal to 1.
complementaryWeight = 1.0 - sum(listOfWeights)
weights = list(listOfWeights)
weights.append(complementaryWeight)
The easy part comes: the unique realizations can be used to fit a kernel smoothing. The KDE is then added to the list of atoms.
sampleUniques = ot.Sample(uniqueValues, 1)
factory = ot.KernelSmoothing()
kde = factory.build(sampleUniques)
atoms = list(listOfDiracs)
atoms.append(kde)
Et voilà: the Mixture is ready.
mixture_estimated = ot.Mixture(atoms, weights)
The following script compares the initial Mixture and the estimated one.
graph = DrawMixtureWithDiracs(distribution)
graph.setColors(["dodgerblue3", "dodgerblue3"])
curve = DrawMixtureWithDiracs(mixture_estimated)
curve.setColors(["darkorange1", "darkorange1"])
curve.setLegends(["Est. Mixture", "Est. Dirac"])
graph.add(curve)
graph
The figure seems satisfactory, since the continuous distribution is estimated from a sub-sample which size is only equal to 50, i.e. one half of the full sample.