I am trying to design a schema for the a clinical dataset, MIMIC-III. I am trying to figure out a way to store the events in a way such that users can query the dataset with ease using possibly a star schema. Almost all of the entries such as diagnoses, procedures, notes, charts etc. are related to a single admission. I had a few things in my mind but I have no experience so am finding it difficult to figure out what the best way to do this is.
- Create multiple fact tables, for example I would have one fact table for diagnoses, one for procedures, one for lab notes but this just seems like too many fact tables with little to gain. Like I could have a fact table with an entry for each diagnosis per user per admission but would that give me more benefit than the OLTP schema already implemented?
- Create one fact table with a row per admission with multiple columns/dimensions like diagnoses, procedures etc. But the issue with is that for most there are multiple diagnoses per admission so I will have to link to a bridge table in most fact tables and then it would look like the image below. The issue with this is the required joins for the queries.
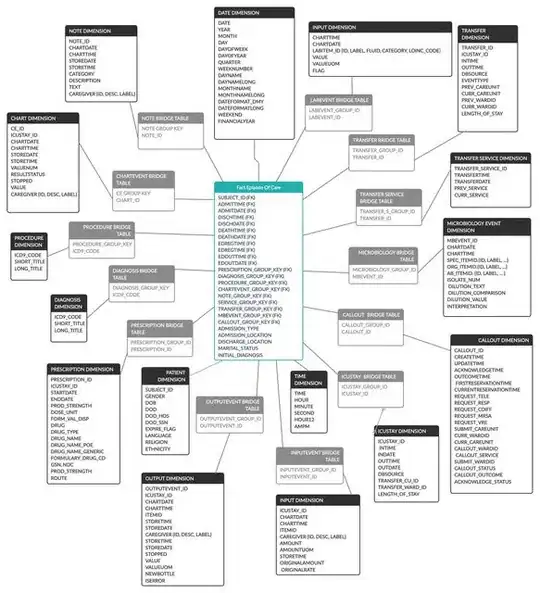
- The third way that I read about is using something like an entity-attribute-value fact table where I have a fact table with each row being one fact. For example, one row could be something like (patientid - addmissionid - (Attribute) Heart Rate Reading - (VALUE) 120bpm) this would create a single fact table with almost everything inside and fewer joins required but it would require the end user to pivot the table after querying due to the nature of EAV's thus making it more complicated for the end user. It would look like the i2b2 star schema.
- The last way I thought of was doing an entry per event into the fact table but having many columns in the fact table to store dimensions, like (patientid, admissionid, icustay_id, diagnosis, procedure, labnote, labevent, microbiologyevent, etc.) in which patientid, and admissionid will be in all rows but the rest will depend on the entry so one entry could have just patientid, admissionid and a single procedure. I don't know how the end result of this will be like in terms of querying due to my lack of experience. I also don't know whether or not all these entries with almost every column being irrelevant is the way to go.
Any help would be greatly appreciated, I'm trying to have this implemented into BigQuery.