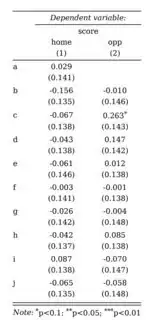
My model output features 32 coefficients. I want them displayed all in one table and came up with the idea to show 16 coeffients in one column respectively with their inherent estimates and p-values. Is there any way to slice the model output or tell stargazer to split the output into different columns?
poisson_model <-
bind_rows(
tibble(
goals = database_mr$goals_team_home,
team = database_mr$club_name_home,
opponent=database_mr$club_name_away,
home=1),
tibble(
goals=database_mr$goals_team_away,
team=database_mr$club_name_away,
opponent=database_mr$club_name_home,
home=0)) %>%
glm(goals ~ home + team +opponent, family=poisson(link=log),data=.)
summary(poisson_model)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.75216 0.22805 3.298 0.000973 ***
home 0.24096 0.07588 3.176 0.001495 **
teamAdler Weseke II -1.04748 0.24868 -4.212 2.53e-05 ***
teamBVH Dorsten -0.28911 0.19946 -1.449 0.147200
teamFC RW Dorsten -0.87653 0.23168 -3.783 0.000155 ***
teamFenerbahce I. Marl -0.56356 0.20580 -2.738 0.006175 **
teamSC Marl-Hamm -0.14523 0.19169 -0.758 0.448688
teamSC Reken II -0.40481 0.20569 -1.968 0.049057 *
teamSV Altendorf-Ulfkotte -1.25184 0.27720 -4.516 6.30e-06 ***
teamSV Lembeck -0.21607 0.19568 -1.104 0.269518
teamSV Schermbeck II -0.16674 0.18600 -0.896 0.370028
teamTSV Raesfeld 0.02094 0.17866 0.117 0.906682
teamTuS 05 Sinsen II -0.90159 0.24070 -3.746 0.000180 ***
teamTuS Gahlen -0.26630 0.19142 -1.391 0.164171
teamTuS Velen -0.40946 0.20151 -2.032 0.042159 *
teamVfL Ramsdorf 0.07215 0.17726 0.407 0.683973
teamWestfalia Gemen II -0.55929 0.20990 -2.665 0.007709 **
opponentAdler Weseke II 0.59518 0.21831 2.726 0.006405 **
opponentBVH Dorsten 0.05072 0.25027 0.203 0.839389
opponentFC RW Dorsten 0.17760 0.23700 0.749 0.453647
opponentFenerbahce I. Marl 0.10922 0.24428 0.447 0.654802
opponentSC Marl-Hamm 0.50746 0.22592 2.246 0.024691 *
opponentSC Reken II 0.69698 0.21994 3.169 0.001530 **
opponentSV Altendorf-Ulfkotte 1.08930 0.20466 5.322 1.02e-07 ***
opponentSV Lembeck 0.35564 0.22962 1.549 0.121428
opponentSV Schermbeck II -0.26666 0.27163 -0.982 0.326254
opponentTSV Raesfeld -0.08465 0.25771 -0.328 0.742563
opponentTuS 05 Sinsen II 0.58102 0.21870 2.657 0.007891 **
opponentTuS Gahlen -0.81158 0.31450 -2.581 0.009865 **
opponentTuS Velen 0.28034 0.23333 1.201 0.229578
opponentVfL Ramsdorf -0.43481 0.28270 -1.538 0.124030
opponentWestfalia Gemen II 0.59072 0.22016 2.683 0.007293 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1