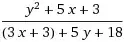I am extending the Virus on Network example from Mesa. The current network graph is like this. However, I want to remove the edges from the dead hosts (black).
My attempt is within the try_check_death():
model_agent.py
import random
import pysnooper
import sys
from random import randrange
from mesa import Agent
from .model_state import State
class HostAgent(Agent):
def __init__(self, unique_id, model, initial_state, virus_check_frequency, chance_spread_virus,
chance_recovery, chance_gain_resistance, chance_virus_kill_host, chance_severe_condition):
super().__init__(unique_id, model)
self.age = randrange(101) # Later to be reassign by another module (AB, Edmonton, Calgary specific)
self.sex = random.choice(['M', 'F']) # Later to be reassign by another module (AB, Edmonton, Calgary specific)
self.urban = random.choice(['Urban', 'Rural']) # Later to be reassign by another module (AB, Edmonton, Calgary specific); later to be interact with agent location and link assignment
self.state = initial_state
self.virus_check_frequency = virus_check_frequency
self.chance_spread_virus = chance_spread_virus
self.chance_recovery = chance_recovery
self.chance_gain_resistance = chance_gain_resistance
self.chance_virus_kill_host = chance_virus_kill_host
self.chance_severe_condition = chance_severe_condition
self.days_of_infection = None
self.days_in_hospital_for_infection = None
self.infection_severity = None
self.clinical_outcomes_from_infection = None
self.clinical_outcomes_after_recovery = None
def modify_chance(self): # a generic function that modify chance attributes
pass
def try_infect_neighbors(self):
neighbors_nodes = self.model.grid.get_neighbors(self.pos, include_center=False)
susceptible_neighbors = [agent for agent in self.model.grid.get_cell_list_contents(neighbors_nodes) if
agent.state is State.SUSCEPTIBLE]
for neighbor_agent in susceptible_neighbors:
if self.random.random() < self.chance_spread_virus:
neighbor_agent.state = State.INFECTED
def try_gain_resistance(self):
if self.random.random() < self.chance_gain_resistance:
self.state = State.RESISTANT
def try_remove_infection(self):
# Try to remove
if self.random.random() < self.chance_recovery:
# Success
self.state = State.SUSCEPTIBLE
self.try_gain_resistance()
else:
# Failed
self.state = State.INFECTED
def try_kill_host(self):
if self.random.random() < self.chance_virus_kill_host:
self.state = State.DEATH
def try_check_infection(self):
if self.random.random() < self.virus_check_frequency:
# Checking...
if self.state is State.INFECTED:
self.try_remove_infection()
def try_check_death(self):
if self.state is State.INFECTED:
self.try_kill_host()
if self.state is State.DEATH:
neighbors_nodes = self.model.grid.get_neighbors(self.pos, include_center=False)
neighbor_agents = [neighbor for neighbor in self.model.grid.get_cell_list_contents(neighbors_nodes)]
agent_neighbor_pairs = [(self.unique_id, neighbor.unique_id) for neighbor in neighbor_agents]
self.model.G.remove_edges_from(agent_neighbor_pairs)
def step(self):
if self.state is State.INFECTED:
self.try_infect_neighbors()
self.try_check_death()
self.try_check_infection()
model_network.py
import math
import sys
import networkx as nx
from mesa import Model
from mesa.time import RandomActivation
from mesa.datacollection import DataCollector
from mesa.space import NetworkGrid
from .model_state import State, number_infected, number_susceptible, number_resistant, number_state, number_death
from .model_agent import HostAgent
class HostNetwork(Model):
"""A virus model with some number of agents"""
def __init__(self, num_nodes=0, avg_node_degree=0, initial_outbreak_size=1, chance_spread_virus=0.0,
virus_check_frequency=0.0, chance_recovery=0.0, chance_gain_resistance=0.0,
chance_virus_kill_host=0.1, chance_severe_condition=0.0):
# Some assert statement to make sure some chances together don't add up > 1.0
self.num_nodes = num_nodes
prob = avg_node_degree / self.num_nodes
self.G = nx.erdos_renyi_graph(n=self.num_nodes, p=prob)
self.grid = NetworkGrid(self.G)
self.schedule = RandomActivation(self)
self.initial_outbreak_size = initial_outbreak_size if initial_outbreak_size <= num_nodes else num_nodes
self.chance_spread_virus = chance_spread_virus
self.virus_check_frequency = virus_check_frequency
self.chance_recovery = chance_recovery
self.chance_gain_resistance = chance_gain_resistance
self.chance_virus_kill_host = chance_virus_kill_host
self.chance_severe_condition = chance_severe_condition
self.datacollector = DataCollector({"Infected": number_infected,
"Susceptible": number_susceptible,
"Resistant": number_resistant,
"Death": number_death,
})
# Create agents
for i, node in enumerate(self.G.nodes()):
agent = HostAgent(i, self, State.SUSCEPTIBLE, self.chance_spread_virus, self.virus_check_frequency,
self.chance_recovery, self.chance_gain_resistance, self.chance_virus_kill_host,
self.chance_severe_condition)
self.schedule.add(agent)
# Add the agent to the node
self.grid.place_agent(agent, node)
# Infect some nodes
infected_nodes = self.random.sample(self.G.nodes(), self.initial_outbreak_size)
for agent in self.grid.get_cell_list_contents(infected_nodes):
agent.state = State.INFECTED
self.running = True
self.datacollector.collect(self)
def resistant_susceptible_ratio(self):
try:
return number_state(self, State.RESISTANT) / number_state(self, State.SUSCEPTIBLE)
except ZeroDivisionError:
return math.inf
def step(self):
self.schedule.step()
# collect data
self.datacollector.collect(self)
def run_model(self, n):
for i in range(n):
self.step()
server.py
import sys
import math
from mesa.visualization.ModularVisualization import ModularServer
from mesa.visualization.UserParam import UserSettableParameter
from mesa.visualization.modules import ChartModule
from mesa.visualization.modules import NetworkModule
from mesa.visualization.modules import TextElement
from .model_network import HostNetwork
from .model_state import State, number_infected, number_susceptible, number_resistant, number_death
def network_portrayal(G):
# The model ensures there is always 1 agent per node
def node_color(agent):
return {
State.INFECTED: '#FF0000',
State.SUSCEPTIBLE: '#008000',
State.DEATH: '#000000',
}.get(agent.state, '#00C5CD')
def edge_color(agent1, agent2):
if State.RESISTANT in (agent1.state, agent2.state):
return '#000000'
return '#e8e8e8'
def edge_width(agent1, agent2):
if State.RESISTANT in (agent1.state, agent2.state):
return 1
else:
return 3
def get_agents(source, target):
return G.nodes[source]['agent'][0], G.nodes[target]['agent'][0]
portrayal = dict()
portrayal['nodes'] = [{'size': 6,
'color': node_color(agents[0]),
'tooltip': "id: {}<br>state: {}".format(agents[0].unique_id, agents[0].state.name),
}
for (_, agents) in G.nodes.data('agent')]
portrayal['edges'] = [{'source': source,
'target': target,
'color': edge_color(*get_agents(source, target)),
'width': edge_width(*get_agents(source, target)),
}
for (source, target) in G.edges]
return portrayal
network = NetworkModule(network_portrayal, 500, 500, library='d3')
chart = ChartModule([{'Label': 'Infected', 'Color': '#FF0000'},
{'Label': 'Susceptible', 'Color': '#008000'},
{'Label': 'Resistant', 'Color': '#00C5CD'},
{'Label': 'Death', 'Color': '#000000'},
])
class MyTextElement(TextElement):
def render(self, model):
ratio = model.resistant_susceptible_ratio()
resistance_susceptible_ratio_text = '∞' if ratio is math.inf else '{0:.2f}'.format(ratio)
infected_text = str(number_infected(model))
susceptible_text = str(number_susceptible(model))
resistant_text = str(number_resistant(model))
death_text = str(number_death(model))
return "Resistant/Susceptible Ratio: {}\
<br>Infected Number: {}\
<br>Susceptible Number: {}\
<br>Resistant Number: {}\
<br>Death Number: {}\
".format(resistance_susceptible_ratio_text, infected_text, susceptible_text,
resistant_text, death_text)
model_params = {
'num_nodes': UserSettableParameter('slider', 'Number of agents', 10, 10, 300, 1,
description='Choose how many agents to include in the model'),
'avg_node_degree': UserSettableParameter('slider', 'Avg Node Degree', 2, 1, 8, 1,
description='Avg Node Degree'),
'initial_outbreak_size': UserSettableParameter('slider', 'Initial Outbreak Size', 1, 1, 100, 1,
description='Initial Outbreak Size'),
'chance_spread_virus': UserSettableParameter('slider', 'Chance to spread virus', 0.4, 0.0, 1.0, 0.1,
description='Probability that susceptible neighbor will be infected'),
'virus_check_frequency': UserSettableParameter('slider', 'Virus Check Frequency', 0.4, 0.0, 1.0, 0.1,
description='Frequency the nodes check whether they are infected by '
'a virus'),
'chance_recovery': UserSettableParameter('slider', 'Chance to recover', 0.3, 0.0, 1.0, 0.1,
description='Probability that the virus will be removed'),
'chance_gain_resistance': UserSettableParameter('slider', 'Chance to gain resistance', 0.5, 0.0, 1.0, 0.1,
description='Probability that a recovered agent will become '
'resistant to this virus in the future'),
}
server = ModularServer(HostNetwork, [network, MyTextElement(), chart], 'Covid-19 Model', model_params)
server.port = 8521
self.model.G.remove_edges_from(agent_neighbor_pairs) seems to give me troubles. What I am seeing is some duplicated nodes and edges similar (but not exactly the same) as the original graph.