The broader question I am interested in is how to interpret differently doing a PERMANOVA on an species-abundance table vs. on its distance matrix (I use bray-curtis).
On the R documentation on the adonis function (https://rdrr.io/rforge/vegan/man/adonis.html), one clue I found is that they mention that one difference is that "species coefficients" cannot be calculated on the distance matrix.
My questions are:
- What are "species coefficients"?
- How to get them?
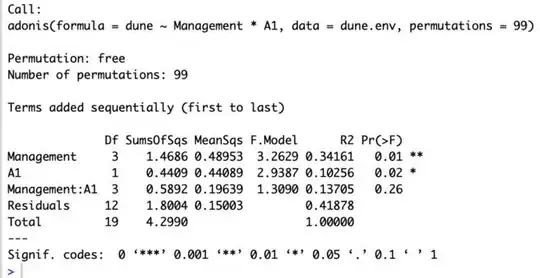
Example: below are the commands and the output I get (using the example in the documentation). They mention that the coefficients should show sources of variations as rows, and species as column headers, and columns the fit to the linear model. I don't see it in the output, how do I get it? How do you interpret these coefficients?
Commands:
data(dune)
data(dune.env)
adonis(dune ~ Management*A1, data=dune.env, permutations=99)
Bonus question: do you have other ways to explain why you would perform the PERMANOVA on the distance matrix rather than on the species-abundance table?