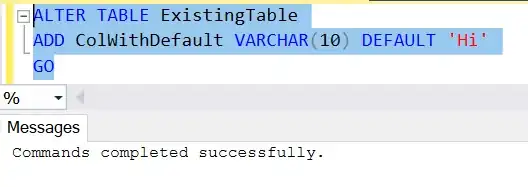
I had to apply Levenshtein Function on last column when passport and country are same.
matrix = passport_heck.select(\
f.col('name_id').alias('name_id_1'),
f.col('last').alias('last_1'),
f.col('country').alias('country_1'),
f.col('passport').alias('passport_1')) \
.crossJoin(passport_heck.select(\
f.col('name_id').alias('name_id_2'),
f.col('last').alias('last_2'),
f.col('country').alias('country_2'),
f.col('passport').alias('passport_2')))\
.filter((f.col('passport_1') == f.col('passport_2')) & (f.col('country_1') == f.col('country_2')))```
res = matrix.withColumn('distance', levenshtein(f.col('last_1'), f.col('last_2')))
Now I am getting the following output which is totally fine.
Now I need to delete duplicates pair (example ID 558635 with 1106562 then 1106562 with 558635 comparing same content).
Can anyone please give me some logic in pyspark to get below table.