I acquired data (motor adaptation =y in function of delays =t ) which I expect to look like a sine wave. I am trying to:
- Fit a sine curve in my data
- Estimate the best model/parameters.
I have read several posts here, here and here but I am still struggling.
1) Using lm
Code:
t<-c(0, 1, 2, 3, 4, 5, 6, 7, 8, 9)
y<-c(0.310 ,0.630 ,0.430 ,0.245, 0.650 ,0.085 ,0.370, 0.560 ,0.250, 0.520)
reslm <- lm(y ~ sin(pi/2*t)+ cos(pi/2*t)) #my period is supposed to be 4, so period equals to pi/2
summary(reslm)
rg<-(max(y)-min(y)/2)
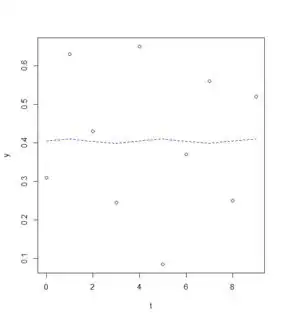
plot(y~t)
lines(fitted(reslm)~t,col=4,lty=2)
Output:
lm(formula = y ~ sin(pi/2 * t) + cos(pi/2 * t))
Residuals:
Min 1Q Median 3Q Max
-0.32450 -0.13956 -0.00325 0.14819 0.24450
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.404375 0.067993 5.947 0.000572 ***
sin(pi/2 * t) 0.005125 0.095190 0.054 0.958567
cos(pi/2 * t) 0.001125 0.095190 0.012 0.990900
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.2107 on 7 degrees of freedom
Multiple R-squared: 0.0004303, Adjusted R-squared: -0.2852
F-statistic: 0.001507 on 2 and 7 DF, p-value: 0.998
Graph:
Questions:
- I am super confused, how can I change the amplitude as well as the phase shift?
- How can I improve my fit using this methods?
2) Using nls
I used the equation y(t) = A*sin(Omega*t + Phi) + C where A is the amplitude, Omega the period, Phi the phase shift and C the midline.
Code:
t<-c(0, 1, 2, 3, 4, 5, 6, 7, 8, 9)
y<-c(0.310 ,0.630 ,0.430 ,0.245, 0.650 ,0.085 ,0.370, 0.560 ,0.250, 0.520)
A<- (max(y)-min(y)/2)
C<-((max(y)+min(y))/2)
res1<- nls(y ~ A*sin(omega*t+phi)+C, data=data.frame(t,y), start=list(A=A,omega=pi/2,phi=0,C=C))
summary(res1)
co <- coef(res1)
resid(res1)
sum(resid(res1)^2)
fit <- function(x, a, b, c, d) {a*sin(b*x+c)+d}
# Plot result
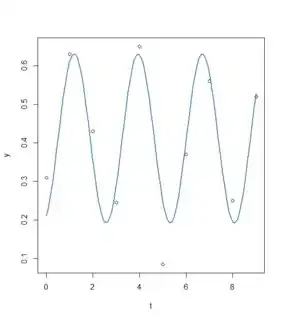
plot(x=t, y=y)
curve(fit(x, a=co["A"], b=co["omega"], c=co["phi"], d=co["C"]), add=TRUE ,lwd=2, col="steelblue")
Output:
Formula: y ~ A * sin(omega * t + phi) + C
Parameters:
Estimate Std. Error t value Pr(>|t|)
A 0.21956 0.03982 5.513 0.0015 **
omega 2.28525 0.07410 30.841 7.72e-08 ***
phi -32.57364 0.40375 -80.678 2.44e-10 ***
C 0.41146 0.02926 14.061 8.07e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.09145 on 6 degrees of freedom
Number of iterations to convergence: 18
Achieved convergence tolerance: 9.705e-06
Graph:
Questions:
- This methods seem to work a bit better, but how can I improve the fit using this method? I tried to change some parameters manually, for example, changing the phase shift (phi) does not do much or lead to an error (see part 3).
3) Using nls and nsl2, in order to tune my model
Code:
###nls2
t<-c(0, 1, 2, 3, 4, 5, 6, 7, 8, 9)
y<-c(0.310 ,0.630 ,0.430 ,0.245, 0.650 ,0.085 ,0.370, 0.560 ,0.250, 0.520)
A <- (max(y)-min(y)/2)
C<-((max(y)+min(y))/2)
pp <- expand.grid(omega=(c(2.094395, 1.570796, 1.256637)), phi=(-1:1), A=A, C=C) # omega = 2*pi/3, pi/2 , 2*pi/5
View(pp)
pp1<-data.frame(pp)
res2<- nls2(y ~ A*sin(omega*t+phi)+C, data=data.frame(t,y), start=pp1, algorithm = "brute-force")
res2
summary(res2)
co <- coef(res2)
resid(res2)
sum(resid(res2)^2)
fit <- function(x, a, b, c, d) {a*sin(b*x+c)+d}
# Plot result
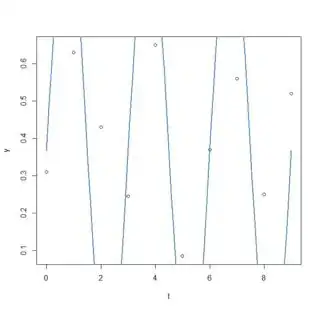
plot(x=t, y=y)
curve(fit(x, a=co["A"], b=co["omega"], c=co["phi"], d=co["C"]), add=TRUE ,lwd=2, col="steelblue")
#optimisation
res3<-nls2(y ~ A*sin(omega*t+phi)+C, start = res2)
res3
summary(res3)
co3 <- coef(res3)
resid(res3)
sum(resid(res3)^2)
fit <- function(x, a, b, c, d) {a*sin(b*x+c)+d}
# Plot result
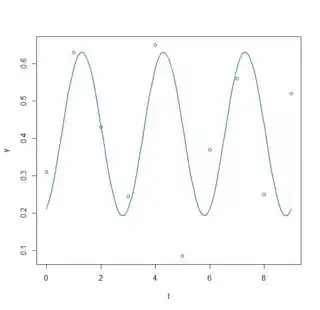
plot(x=t, y=y)
curve(fit(x, a=co3["A"], b=co["omega"], c=co3["phi"], d=co3["C"]), add=TRUE ,lwd=2, col="steelblue")
Output:
First attempt (nls2 model1):
model: y ~ A * sin(omega * t + phi) + C
data: data.frame(t, y)
omega phi A C
2.0944 0.0000 0.6075 0.3675
residual sum-of-squares: 0.8545
Number of iterations to convergence: 9
Achieved convergence tolerance: NA
> summary(res2)
Formula: y ~ A * sin(omega * t + phi) + C
Parameters:
Estimate Std. Error t value Pr(>|t|)
omega 2.09440 0.08453 24.776 2.84e-07 ***
phi 0.00000 0.46494 0.000 1.0000
A 0.60750 0.17851 3.403 0.0144 *
C 0.36750 0.12044 3.051 0.0225 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.3774 on 6 degrees of freedom
Number of iterations to convergence: 9
Achieved convergence tolerance: NA
Graph:
Second attempt (nls2 model2):
Nonlinear regression model
model: y ~ A * sin(omega * t + phi) + C
data: <environment>
omega phi A C
2.2852 -1.1577 0.2196 0.4115
residual sum-of-squares: 0.05018
Number of iterations to convergence: 12
Achieved convergence tolerance: 8.075e-06
> summary(res3)
Formula: y ~ A * sin(omega * t + phi) + C
Parameters:
Estimate Std. Error t value Pr(>|t|)
omega 2.28524 0.07410 30.841 7.72e-08 ***
phi -1.15769 0.40375 -2.867 0.0285 *
A 0.21956 0.03982 5.513 0.0015 **
C 0.41146 0.02926 14.061 8.07e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.09145 on 6 degrees of freedom
Number of iterations to convergence: 12
Achieved convergence tolerance: 8.075e-06
Graph:
Questions:
- So here it seems I misunderstood what nls2 was doing as I am finding exactly the same results as part 2. I still do not know which parameters are the best, how can I do this?
4) Using nls, in order to tune my model by looping through several parameters
Code:
t<-c(0, 1, 2, 3, 4, 5, 6, 7, 8, 9)
y<-c(0.310 ,0.630 ,0.430 ,0.245, 0.650 ,0.085 ,0.370, 0.560 ,0.250, 0.520)
A <- (max(y)-min(y)/2)
C<-((max(y)+min(y))/2)
pp <- expand.grid(omega=(c(2.094395, 1.570796, 1.256637)), phi=(-1:1), A=A, C=C) # omega = 2*pi/3, pi/2 , 2*pi/5
#View(pp)
fit_AIC<- vector()
fit_BIC<- vector()
coef_A<- vector()
coef_ome<- vector()
coef_phi<- vector()
coef_C<- vector()
RSS<-vector()
for (ii in 1:nrow(pp))
{
res<- nls(y ~ A*sin(omega*t+phi)+C, data=data.frame(t,y), start=list(A=pp$A[ii],omega=pp$omega[ii],phi=pp$phi[ii],C=pp$C[ii]), trace = TRUE)
fit_AIC[ii]<-AIC(res)
fit_BIC[ii]<-BIC(res)
coef_A[ii]<- coef(res)[1]
coef_ome[ii]<- coef(res)[2]
coef_phi[ii]<- coef(res)[3]
coef_C[ii]<- coef(res)[4]
RSS<-sum(resid(res)^2)
}
results<-data.frame(RSS, fit_AIC, fit_BIC, coef_A, coef_ome, coef_phi, coef_C)
View(results)
Output:
I get this error:
1.405742 : 0.607500 2.094395 -1.000000 0.367500
0.1448148 : 0.1563179 2.1441802 -0.9937729 0.4172079
...
0.05018035 : 0.2195573 2.2852482 -1.1577097 0.4114573
2.085664 : 0.607500 1.570796 1.000000 0.367500
0.3104012 : 0.01321257 1.60518024 0.83201816 0.40437498
0.3098916 : 0.0180852 3.0888764 -5.9933691 0.4060743
Error in nls(y ~ A * sin(omega * t + phi) + C, data = data.frame(t, y), :
le pas 0.000488281 est devenu inférieur à 'minFactor' de 0.000976562
RSS fit_AIC fit_BIC coef_A coef_ome coef_phi coef_C
1 0.05018035 -14.568398 -13.055473 0.21955754 2.2852455 -1.1576955 0.4114573
2 0.05018035 2.753153 4.266079 0.07487110 0.8575642 0.2299909 0.3916769
3 0.05018035 2.753153 4.266079 0.07487109 0.8575736 0.2299951 0.3916763
4 0.05018035 -14.568398 -13.055473 0.21955763 2.2852443 -1.1576894 0.4114573
5 0.05018035 -14.568398 -13.055473 0.21955729 2.2852490 -32.5736406 0.4114573
6 0.05018035 2.753153 4.266079 0.07487105 0.8575619 0.2300021 0.3916770
7 0.05018035 -14.568398 -13.055473 0.21955735 2.2852482 -1.1577097 0.4114573
Questions:
- So this error seems to be because my initial parameters are wrong. Is this correct? But how can I estimate the best parameters if the majority of parameters do not work?
- Also, I do not understand why the RSS is always the same despite different parameters
- And why do I observe only 2 different AIC and 2 different BIC while the models are different?
Any kind of help would be much appreciated, thanks.