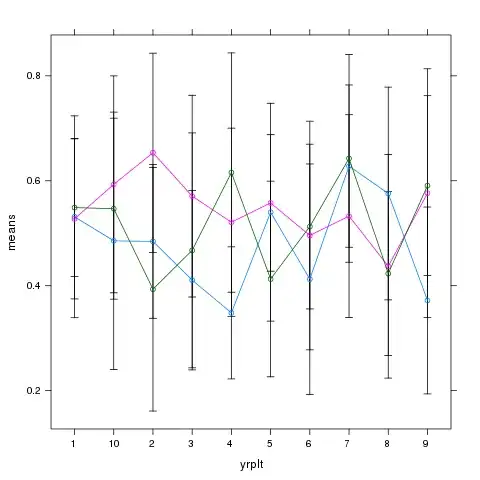
I calculated a linear-mixed model using the nlme package. I was evaluating a psychological treatment and used treatment condition and measurement point as predictors. I did post-hoc comparisons using the emmans package. So far so good, everything worked out well and I am looking forward to finish my thesis. There is only one problem left. I am really really bad in plotting. I want to plot the emmeans for the four measurement points for each group. The emmip function in emmeans does this, but I am not that happy with the result. I used the following code to generate the result:
emmip(HLM_IPANAT_pos, Gruppe~TP, CIs=TRUE) + theme_bw() + labs(x = "Zeit", y = "IPANAT-PA")
I don't like the way the confidence intervals are presented. I would prefer a line bar with "normal" confidence bars, like the one below, which is taken from Ireland et al. (2017). I tried to do it in excel, but did not find out how to integrate seperate confidence intervals for each line. So I was wondering if there was the possibility to do it using ggplot2. However, I do not know how to integrate the values I obtained using emmeans in ggplot. As I said, I really have no idea about plotting. Does someone know how to do it?