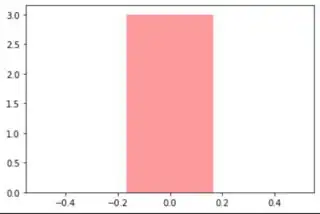
I am trying to create histograms for feature analysis to see how similar high grade tumor and low grade tumor characteristics are. Something similar to the histogram shown below:
Matrix "Z" looks like this and contains 106 features
Matrix "Z"
The code I am using is:
#Import data
data = pd.read_csv("/Users/MLuser/Desktop/Data table - CM_Transposed.csv")
# Prproccess data
bins = (0,2,4)
group_names = ['low_grade','high_grade']
data['FGrade'] = pd.cut(data['FGrade'], bins=bins, labels=group_names)
label_grade = LabelEncoder()
data['FGrade'] = label_grade.fit_transform(data['FGrade'])
Z=data
# Separate the dataset as response variable and feature variables
Z = Z.drop('Feature Name', axis = 1)
Z.columns
low_grade=Z[Z.FGrade==0] # define malignant
high_grade=Z[Z.FGrade==1] # define benign
for i, col in enumerate(Z.columns):
plt.figure(i)
sns.distplot(low_grade.iloc[:,i], color='red')
sns.distplot(high_grade.iloc[:,i], color='g')
Unfortunately when I run the above, I get a single graph and the following error:
---------------------------------------------------------------------------
LinAlgError Traceback (most recent call last)
<ipython-input-25-79d6e8cd51a8> in <module>
27 for i, col in enumerate(Z.columns):
28 plt.figure(i)
---> 29 sns.distplot(low_grade.iloc[:,i], color='red')
30 sns.distplot(high_grade.iloc[:,i], color='g')
31
~/env/lib/python3.6/site-packages/seaborn/distributions.py in distplot(a, bins, hist, kde, rug, fit, hist_kws, kde_kws, rug_kws, fit_kws, color, vertical, norm_hist, axlabel, label, ax)
229 if kde:
230 kde_color = kde_kws.pop("color", color)
--> 231 kdeplot(a, vertical=vertical, ax=ax, color=kde_color, **kde_kws)
232 if kde_color != color:
233 kde_kws["color"] = kde_color
~/env/lib/python3.6/site-packages/seaborn/distributions.py in kdeplot(data, data2, shade, vertical, kernel, bw, gridsize, cut, clip, legend, cumulative, shade_lowest, cbar, cbar_ax, cbar_kws, ax, **kwargs)
689 ax = _univariate_kdeplot(data, shade, vertical, kernel, bw,
690 gridsize, cut, clip, legend, ax,
--> 691 cumulative=cumulative, **kwargs)
692
693 return ax
~/env/lib/python3.6/site-packages/seaborn/distributions.py in _univariate_kdeplot(data, shade, vertical, kernel, bw, gridsize, cut, clip, legend, ax, cumulative, **kwargs)
292 "only implemented in statsmodels."
293 "Please install statsmodels.")
--> 294 x, y = _scipy_univariate_kde(data, bw, gridsize, cut, clip)
295
296 # Make sure the density is nonnegative
~/env/lib/python3.6/site-packages/seaborn/distributions.py in _scipy_univariate_kde(data, bw, gridsize, cut, clip)
364 """Compute a univariate kernel density estimate using scipy."""
365 try:
--> 366 kde = stats.gaussian_kde(data, bw_method=bw)
367 except TypeError:
368 kde = stats.gaussian_kde(data)
~/env/lib/python3.6/site-packages/scipy/stats/kde.py in __init__(self, dataset, bw_method, weights)
206 self._neff = 1/sum(self._weights**2)
207
--> 208 self.set_bandwidth(bw_method=bw_method)
209
210 def evaluate(self, points):
~/env/lib/python3.6/site-packages/scipy/stats/kde.py in set_bandwidth(self, bw_method)
552 raise ValueError(msg)
553
--> 554 self._compute_covariance()
555
556 def _compute_covariance(self):
~/env/lib/python3.6/site-packages/scipy/stats/kde.py in _compute_covariance(self)
564 bias=False,
565 aweights=self.weights))
--> 566 self._data_inv_cov = linalg.inv(self._data_covariance)
567
568 self.covariance = self._data_covariance * self.factor**2
~/env/lib/python3.6/site-packages/scipy/linalg/basic.py in inv(a, overwrite_a, check_finite)
972 inv_a, info = getri(lu, piv, lwork=lwork, overwrite_lu=1)
973 if info > 0:
--> 974 raise LinAlgError("singular matrix")
975 if info < 0:
976 raise ValueError('illegal value in %d-th argument of internal '
LinAlgError: singular matrix
- OS: 10.14.6
- Python: 3.6.8.final.0
- pd: 0.25.1
- np: 1.17.2
- sns: 0.9.0