I have to calculate a non-linear least-square regression for my ~30 data points following the formula
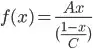
I tried the curve_fit function out of scipy.optimize using the following code
def func(x, p1 ,p2):
return p1*x/(1-x/p2)
popt, pcov = curve_fit(func, CSV[:,1], CSV[:,0])
p1 = popt[0]
p2 = popt[1]
with p1 and p2 being equivalent to A and C, respectively, and CSV being my data-array. The functions runs without error message, but the outcome is not as expected. I've plotted the outcome of the function together with the original data points. I was not looking to get this nearly straight line (red line in plot), but something more close to the green line, which is simply a second order polynomial fit from Excel. The green dashed line shows just a quick manual try to get closer to the polynomial fit.
wrong calcualtin of the fit-function, together with the original data points: 1
Does anyone has an idea how to make the calculation run as i want it to?