It is easier to explain what am I trying to do through the following reproducible example:
library(sf)
library(brazilmaps)
library(spdep)
# loading cities map
cities_mg <- brazilmaps::get_brmap(geo = 'City',
geo.filter = list(State = 31),
class = 'sf')
# creating adjacency matrix (neighborhood graph)
nb_mg <- spdep::poly2nb(cities_mg)
# plotting
coords_mg <- st_coordinates(st_centroid(st_geometry(cities_mg)))
par(bg = '#2E2E2E')
plot(nb_mg, coords_mg, pch = 19, cex = .6, lwd = .4,
col = '#00C040', points = T)
This piece of code generates the following plot
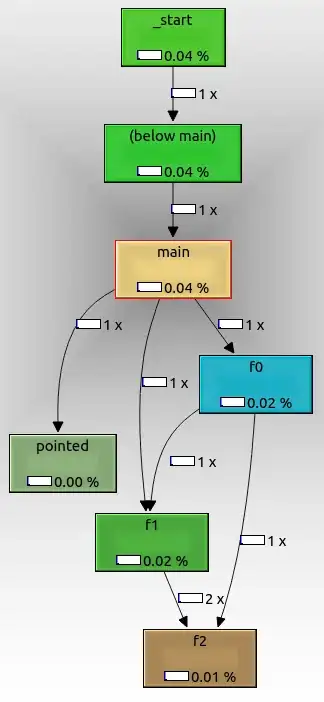
What I would like to do, is coloring the nodes (or vertices) with the same color as the edges. Is it possible?
Thank you in advance.
Session info (for reproducibility) below.
R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 19.04
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.8.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.8.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8
[6] LC_MESSAGES=en_US.UTF-8 LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] spdep_1.1-3 spData_0.3.2 sp_1.3-1 brazilmaps_0.1.0 sf_0.8-0
loaded via a namespace (and not attached):
[1] gtools_3.8.1 tidyselect_0.2.5 xfun_0.10 purrr_0.3.3 splines_3.6.1 lattice_0.20-38 expm_0.999-4
[8] htmltools_0.4.0 XML_3.98-1.20 rlang_0.4.1 pillar_1.4.2 e1071_1.7-2 glue_1.3.1 withr_2.1.2
[15] DBI_1.0.0 semver_0.2.0 binman_0.1.1 caTools_1.17.1.2 coda_0.19-3 evaluate_0.14 knitr_1.25
[22] wdman_0.2.4 callr_3.3.2 ps_1.3.0 class_7.3-15 Rcpp_1.0.2 KernSmooth_2.23-16 clipr_0.7.0
[29] openssl_1.4.1 classInt_0.4-1 gdata_2.18.0 RSelenium_1.7.5 deldir_0.1-23 fs_1.3.1 askpass_1.1
[36] digest_0.6.22 gmodels_2.18.1 processx_3.4.1 dplyr_0.8.3 grid_3.6.1 tools_3.6.1 bitops_1.0-6
[43] LearnBayes_2.15.1 magrittr_1.5 tibble_2.1.3 crayon_1.3.4 pkgconfig_2.0.3 whisker_0.4 MASS_7.3-51.4
[50] Matrix_1.2-17 reprex_0.3.0 rstudioapi_0.10 assertthat_0.2.1 rmarkdown_1.16 R6_2.4.0 boot_1.3-23
[57] units_0.6-5 nlme_3.1-141 compiler_3.6.1