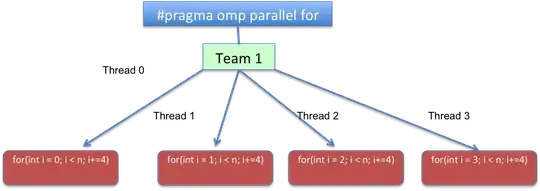
I use prcomp to run PCA in r. When I output the summary, i.e. standard deviation, proportion of variance, cumulative proportion, the results are always ordered and the actual column name is replaced by PC1, PC2. Thus, I cannot tell the exact proportion of variance for each column.
Can anyone show me or give me some hint on how to display the column when outputting summary results. Two results pics are attached here: