Let's say I'm working with proportions, I have two main variables (sex and pain_level). It's not difficult to plot them:
With tidyverse and broom (and thanks for this link here: Calling prop.test function in R with dplyr) I can compare if the proportions are statistically different.
Now comes the question!
I want to add to the plot, the error bar. I know it's not as difficult as I'm thinking, but I could not find a way to do it. I've tried to replicate this link here (http://www.andrew.cmu.edu/user/achoulde/94842/labs/lab07_solution.html) but I'm trying to stay at tidyverse environment.
The desired output should be something like that:
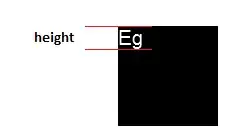
Please feel free to use the script/syntax below that simulate the original dataset.
library(tidyverse)
ds <- data.frame(sex = rep(c("M","F"), 18),
pain_level = c("High","Moderate","low"))
#plot
ds %>%
group_by(pain_level, sex) %>%
summarise(n=n()) %>%
mutate(prop = n/sum(n)*100) %>%
ggplot(., aes(x = sex, fill = pain_level, y = prop)) +
geom_bar(stat = "summary") +
facet_wrap( ~ pain_level) +
theme(legend.position = "none")
#p values of proportion test
ds %>%
rowwise %>%
group_by(pain_level, sex) %>%
summarise(cases = n()) %>%
mutate(pop = sum(cases)) %>% #compute totals
distinct(., pain_level, .keep_all= TRUE) %>% #keep only one value of the row
mutate(tst = list(broom::tidy(prop.test(cases, pop, conf.level=0.95)))) %>%
tidyr::unnest(tst)