So I made my graph using networkx library which is a unipartite graph. When I put the labels it doesn't seem right, its getting mixed up. There are some words which has more length than the others and its out of the boundary of the nodes. Can my graph be adjusted so everything look clear and understandable?
And also I want the node to be like a dot and the labels appearing over the nodes not inside the nodes.
Like this
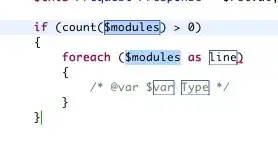
Graph I made..
Here is the code..
import networkx as nx
import matplotlib.pyplot as plt
import numpy as np
from networkx.algorithms import community
from networkx.algorithms import bipartite
G1 = nx.read_edgelist("abcd.txt")
print(nx.info(G1))
c = bipartite.color(G1)
nx.set_node_attributes(G1, c, 'bipartite')
type1 = {n for n, d in G1.nodes(data=True) if d['bipartite']==0}
type2 = {n for n, d in G1.nodes(data=True) if d['bipartite']==1}
G = bipartite.projected_graph(G1, type1)
type2g = bipartite.projected_graph(G1, type2)
pos = nx.spring_layout(G,k=0.30,iterations=50)
nx.is_bipartite(G1)
#average clustering
nx.average_clustering(G1)
#Diameter
print("Diameter:",nx.diameter(G1))
options = {
'node_color': 'purple',
'node_size': 40,
'line_color': 'yellow',
'linewidths': 0,
'width': 0.3,
}
#degeree plotting
def plot_degree_distribution(wiki):
degs = {}
for n in wiki.nodes():
deg = wiki.degree(n)
if deg not in degs:
degs[deg] = 0
degs[deg] += 1
items = sorted(degs.items())
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot([k for (k, v) in items], [v for (k, v) in items])
ax.set_xscale('log')
ax.set_yscale('log')
plt.title("Degree Distribution")
fig.savefig("degree_distribution.png")
# plot_degree_distribution(G)
d = [] # create a set
for n in G.nodes():
d.append(G.degree(n))
ec = [] # create a set
for e in G.edges():
if (G.degree(e[0]) > G.degree(e[1])):
ec.append(G.degree(e[0]))
else:
ec.append(G.degree(e[1]))
pos = nx.spring_layout(G,k=1.5, iterations=200)
factor = 25 # to change the size of nodes with respect to their degree
nx.draw_networkx(G, pos,
edge_color=ec, edge_cmap=plt.cm.plasma, # edge color
node_color=d, cmap=plt.cm.plasma, # node color
node_size=[x * factor for x in d]) # node sizse
plt.savefig ("simple_graph.png")
fig = plt.gcf()
fig.set_size_inches((10,10))
plt.show()