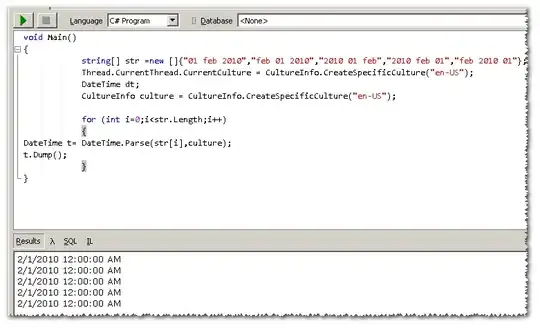
I have a dataframe that I pulled from REDCap, I have imported the CSV in RStudio. unique participant id's are listed and then events. I need to list the number of each repeated event, for example acute_event_infect_arm_4 for 1010002 is the first rejection event and then _4b is the second event, _4c is third and so on.
I need to do this for follow-up, reject and CMV/EBV events
Here is a small snapshot of dataframe with 3 ids (actual df has 1000 ids)
structure(list(id = c(1010002, 1010002, 1010002, 1010002, 1010002,
1010002, 1010002, 1010002, 1010002, 1010002, 1010002, 1010002,
1010002, 1010002, 1010002, 1010002, 1010006, 1010006, 1010006,
1010006, 1010006, 1010006, 1010006, 1010006, 1010006, 1010006,
1010006, 1010008, 1010008, 1010008, 1010008, 1010008, 1010008,
1010008, 1010008, 1010008, 1010008, 1010008), redcap_event_name =
c("pre_transplant_arm_4",
"transplant_arm_4", "transplant_2_arm_4", "end_of_followup_fo_arm_4",
"last_encounter_arm_4", "acute_event_reject_arm_4",
"acute_event_reject_arm_4b",
"acute_event_infect_arm_4", "acute_event_infect_arm_4b",
"acute_event_infect_arm_4c",
"acute_event_infect_arm_4d", "acute_event_infect_arm_4e",
"acute_event_infect_arm_4f",
"acute_event_infect_arm_4g", "acute_event_cmvebv_arm_4",
"acute_event_cmvebv_arm_4b",
"pre_transplant_arm_4", "transplant_arm_4", "1_month_followup_arm_4",
"2_year_followup_arm_4", "last_encounter_arm_4", "acute_event_reject_arm_4",
"acute_event_reject_arm_4b", "acute_event_infect_arm_4",
"acute_event_infect_arm_4b",
"acute_event_infect_arm_4c", "acute_event_cmvebv_arm_4",
"pre_transplant_arm_4",
"transplant_arm_4", "3_month_followup_arm_4", "6_month_followup_arm_4",
"1_year_followup_arm_4", "2_year_followup_arm_4", "3_year_followup_arm_4",
"last_encounter_arm_4", "acute_event_reject_arm_4",
"acute_event_infect_arm_4",
"acute_event_cmvebv_arm_4")), row.names = c(NA, -38L), class = c("tbl_df",
"tbl", "data.frame"))
This is what I need to add in redcap_repeat column
@akrun please some some examples below (bolded red are missing)