I can't get variable labels do not overlap with ggbiplot
(using RStudio 1.1.463 and R version 3.5.3)
I am running a pca with prcomp but i get this kind of variable label overlapping:
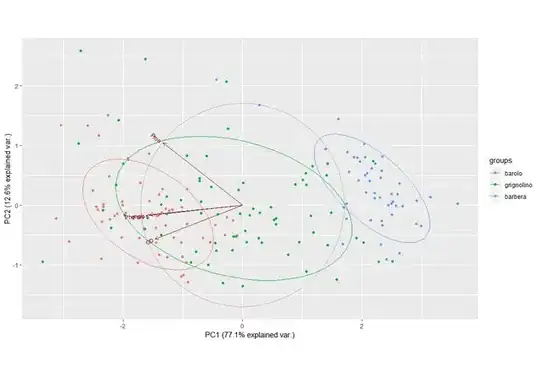
Here is an example:
library(ggbiplot)
data(wine)
wine_subset<-subset(wine[,c(6:7,9,12)])
wine.pca <- prcomp(wine_subset, scale. = TRUE)
print(ggbiplot(wine.pca, obs.scale = 1, var.scale = 1, groups = wine.class, ellipse = TRUE, circle = TRUE))
I tried to solve adding this code from ggrepel package:
library(ggrepel)
+geom_text_repel(aes(labels=colnames(wine_subset)))
but it returns the following error:
Warning: Ignoring unknown aesthetics: labels Error: Aesthetics must be either length 1 or the same as the data (178): labels
It seems to me that it is trying to take the row labels, but I don't need them in the plot. I need the variable labels only.