I would like to scrape drug informations offered by the Swiss government for an University research project from:
http://www.spezialitaetenliste.ch/ShowPreparations.aspx?searchType=Substance&searchValue=
The page does offer a robotx.txt file, however, it's content is freely available to the public and I assume that scraping this data is unprohibited.
This is an update of this question, since I made some progress.
What I achieved so far
# opens the first results page
# opens the first link as a table at the end of the page
library("rvest")
library("dplyr")
url <- "http://www.spezialitaetenliste.ch/ShowPreparations.aspx?searchType=Substance&searchValue="
pgsession<-html_session(url)
pgform<-html_form(pgsession)[[1]]
page<-rvest:::request_POST(pgsession,url,
body=list(
`ctl00$cphContent$gvwPreparations$ctl13$gvwpPreparations$txtPageNumber`=1,
`__VIEWSTATE`=pgform$fields$`__VIEWSTATE`$value,
`__VIEWSTATEGENERATOR`=pgform$fields$`__VIEWSTATEGENERATOR`$value,
`__VIEWSTATEENCRYPTED`=pgform$fields$`__VIEWSTATEENCRYPTED`$value,
`__EVENTVALIDATION`=pgform$fields$`__EVENTVALIDATION`$value,
`ctl00$cphContent$gvwPreparations$ctl13$gvwpPreparations$ddlPageSize`="10",
`__EVENTTARGET`="ctl00$cphContent$gvwPreparations$ctl02$ctl00",
`__EVENTARGUMENT`=""
),
encode="form")
next: get the basic data
# makes a table of all results of the first page
read_html(page) %>%
html_nodes(xpath = '//*[@id="ctl00_cphContent_gvwPreparations"]') %>%
html_table(fill=TRUE) %>%
bind_rows %>%
tibble()
next: get the additional data
# gives the desired informations (=additional data) of the first drug (not yet very structured)
read_html(page) %>%
html_nodes(xpath = '//*[@id="ctl00_cphContent_fvwPreparation"]') %>%
html_text
My Problem:
# if I open the second search page
page<-rvest:::request_POST(pgsession,url,
body=list(
`ctl00$cphContent$gvwPreparations$ctl13$gvwpPreparations$txtPageNumber`=2,
`__VIEWSTATE`=pgform$fields$`__VIEWSTATE`$value,
`__VIEWSTATEGENERATOR`=pgform$fields$`__VIEWSTATEGENERATOR`$value,
`__VIEWSTATEENCRYPTED`=pgform$fields$`__VIEWSTATEENCRYPTED`$value,
`__EVENTVALIDATION`=pgform$fields$`__EVENTVALIDATION`$value,
`ctl00$cphContent$gvwPreparations$ctl13$gvwpPreparations$ddlPageSize`="10",
`__EVENTTARGET`="ctl00$cphContent$gvwPreparations$ctl02$ctl00",
`__EVENTARGUMENT`=""
),
encode="form")
next: get the new basic data
# I get easily a table with the new results
read_html(page) %>%
html_nodes(xpath = '//*[@id="ctl00_cphContent_gvwPreparations"]') %>%
html_table(fill=TRUE) %>%
bind_rows %>%
tibble()
But if I try to get the new additional data, I get the results from page 1 again:
# does not give the desired output:
read_html(page) %>%
html_nodes(xpath = '//*[@id="ctl00_cphContent_fvwPreparation"]') %>%
html_text
What I am looking for: the detailed data of the first drug of page 2
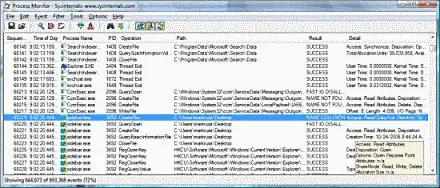
Questions:
- Why do I get duplicate results? Is it because of the
__VIEWSTATEthat might change during the newrequest_POST? - Is there any way to solve this problem?
- Is there any better way how to get the basic and additional data? If yes, how?