I am a gnuplot-newbie and am stuck with the following situation. Based on this I have a gnuplot script as follows:
clear
reset
set key off
set border 3
set style fill solid 1.0 noborder
bin_width = 0.01;
set boxwidth bin_width absolute
bin_number(x) = floor(x/bin_width)
rounded(x) = bin_width * ( bin_number(x) + 0.5 )
plot '1000randomValuesBetween0and1.dat' using (rounded($1)):(1) smooth frequency
Which was a good first step; but I would like to have a smooth curve through the points that are generated by counting the frequency. with filledcurves lacked what I wanted in 2 ways. First it is not smoothed (I would prefer something like bezier which is not usable after with); second the filling is done in a rather unexpected way which doesn't fit my needs (for me unexpected). See this picture 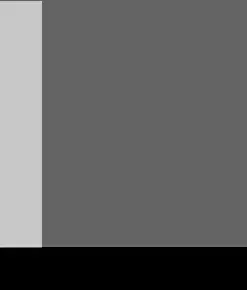 .
.
To give a little bit more context: I ultimately want to use this to generate violin plots with gnuplot without having to do the binning beforehand so I can just give my script a single-column data-file and am ready to go.
EDIT: I tried adapting the "normal" density plot from this demo as another first step, but I failed; I read in the documentation that bandwidth should be 1/#points so it should be 0.001 in my case meaning I tried this:
set border 3 front lt black linewidth 1.000 dashtype solid
set style increment default
set style data filledcurves
set xtics border in scale 0,0 nomirror norotate autojustify
set xtics norangelimit 0.00000,0.5,1.0
set title "Same data - kernel density"
set title font ",15" norotate
plot 'random01.dat' using 1:(1) smooth kdensity bandwidth 0.001 with filledcurves above y lt 9
which results in this picture: .
Setting no bandwith or lower/higher values didn't solve the issue.
The plot specifies using 1:(1) because I just have a single column so according to the doc the first value should be this column and as the second value would specify a weighting which should be 1/#points according to doc.
.
Setting no bandwith or lower/higher values didn't solve the issue.
The plot specifies using 1:(1) because I just have a single column so according to the doc the first value should be this column and as the second value would specify a weighting which should be 1/#points according to doc.
EDIT2: Setting bandwidth to the ideal value or not setting it at all always yields the same result which doesn't change anything except the scale of the y-axis with changing the weighting.
My data are 1000 values in a range between 0 and 1 (created randomly for testing purposes).
EDIT3: zooming out may show another aspect of the problem as the plot seems to extend outside the interval of the given values (I checked the values and there are no examples <0 or >1). Here's the graph: