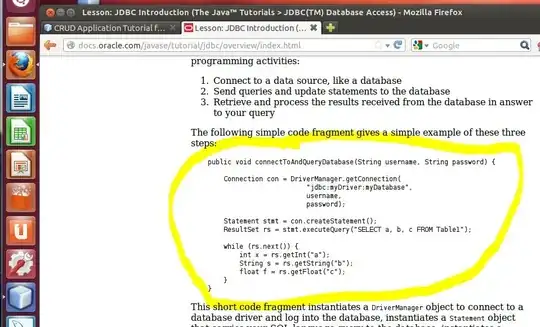
I have a regular square grid wherein all my data points are stored at the centroid. I have a scalar field(range : 0->1) that indicates the amount of substance inside a cell. I am interested in identifying the interface of this substance inside the cell (for further processing and not for visualization).
I came across the Marching cube algorithm(http://paulbourke.net/geometry/polygonise/). Here I need the values at the corners of the cell. So I averaged the centroid values of the neighbouring cell. This averaging coupled with further linearization to find the points of intersection during "Polygonization" in MC is resulting in non-realistic interfaces such as this..
Here the gray cell is full of substance while its neighbours have minimal amount of substance. Ideally this should be very close to the boundary of the celtre cell. I feel this happens due to linear interpolation between 0.25 and 0 which leads it far off from its intended position.
Can something be done to sort out this issue ?