For reproducibility reasons, I am sharing the few datasets here. The dataset has a format of the following.
0.080505471,10
0.080709071,20
0.080835753,30
0.081004589,40
0.081009152,30
0.181258811,41
0.181674244,40
From column 2, I am reading the current row and compare it with the value of the previous row. If it is greater, I keep comparing. If the current value is smaller than the previous row's value, I want to divide the current value (smaller) by the previous value (larger). Accordingly, the following code:
import numpy as np
import scipy.stats
import matplotlib.pyplot as plt
import seaborn as sns
protocols = {}
types = {"data_g": "data_g.csv", "data_v": "data_v.csv", "data_c": "data_c.csv", "data_c": "data_c.csv"}
for protname, fname in types.items():
col_time,col_window = np.loadtxt(fname,delimiter=',').T
trailing_window = col_window[:-1] # "past" values at a given index
leading_window = col_window[1:] # "current values at a given index
decreasing_inds = np.where(leading_window < trailing_window)[0]
quotient = leading_window[decreasing_inds]/trailing_window[decreasing_inds]
quotient_times = col_time[decreasing_inds]
protocols[protname] = {
"col_time": col_time,
"col_window": col_window,
"quotient_times": quotient_times,
"quotient": quotient,
}
plt.figure(); plt.clf()
plt.plot(quotient_times, quotient, ".", label=protname, color="blue")
plt.ylim(0, 1.0001)
plt.title(protname)
plt.xlabel("quotient_times")
plt.ylabel("quotient")
plt.legend()
plt.show()
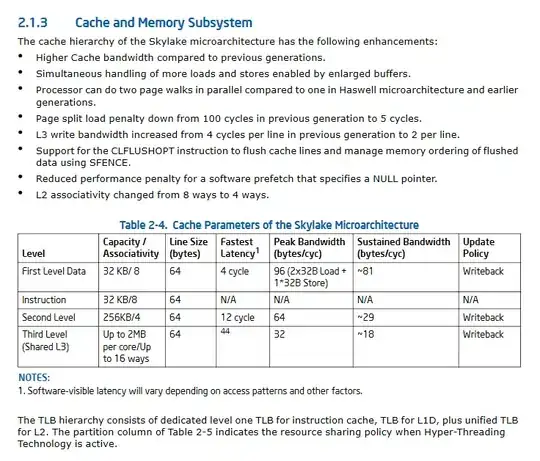
This gives the following plots.
As we can see from the plots
- Data-G, no matter what the value of
quotient_timesis, the quotient is always >=0.9 Data-V has a quotient of 0.8 when the
quotient_timesis less than 3 and the quotient remains 0.5 if thequotient_timesis greater than 3.Data-C has a constant
quotientof 0.7 no matter what the value ofquotient_timesis.Data-R has a constant
quotientof 0.5 no matter what the value ofquotient_times
Based on this requirement, how can we plot a Gaussian Mixture Model? Any help would be appreciated.