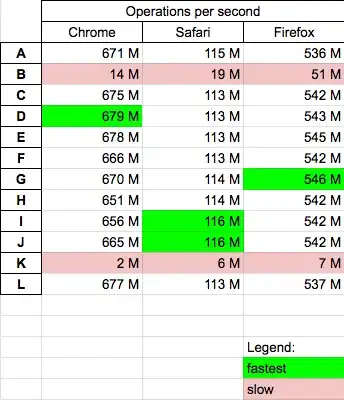
After training an LDA model on gensim LDA model i converted the model to a with the gensim mallet via the malletmodel2ldamodel function provided with the wrapper. Before and after the conversion the topic word distributions are quite different. The mallet version returns very rare topic word distribution after conversion.
ldamallet = gensim.models.wrappers.LdaMallet(mallet_path, corpus=corpus, num_topics=13, id2word=dictionary)
model = gensim.models.wrappers.ldamallet.malletmodel2ldamodel(ldamallet)
model.save('ldamallet.gensim')
dictionary = gensim.corpora.Dictionary.load('dictionary.gensim')
corpus = pickle.load(open('corpus.pkl', 'rb'))
lda_mallet = gensim.models.wrappers.LdaMallet.load('ldamallet.gensim')
import pyLDAvis.gensim
lda_display = pyLDAvis.gensim.prepare(lda_mallet, corpus, dictionary, sort_topics=False)
pyLDAvis.display(lda_display)
Here is the output from gensim original implementation:
I can see there was a bug around this issue which has been fixed with the previous versions of gensim. I am using gensim=3.7.1