I have recently hit a roadblock when it comes to performance. I know how to manually loop and do the interpolation from the origin cell to all the other cells by brute-forcing/looping each row and column in 2d array.
however when I process a 2D array of a shape say (3000, 3000), the linear spacing and the interpolation come to a standstill and severely hurt performance.
I am looking for a way I can optimize this loop, I am aware of vectorization and broadcasting just not sure how I can apply it in this situation.
I will explain it with code and figures
import numpy as np
from scipy.ndimage import map_coordinates
m = np.array([
[10,10,10,10,10,10],
[9,9,9,10,9,9],
[9,8,9,10,8,9],
[9,7,8,0,8,9],
[8,7,7,8,8,9],
[5,6,7,7,6,7]])
origin_row = 3
origin_col = 3
m_max = np.zeros(m.shape)
m_dist = np.zeros(m.shape)
rows, cols = m.shape
for col in range(cols):
for row in range(rows):
# Get spacing linear interpolation
x_plot = np.linspace(col, origin_col, 5)
y_plot = np.linspace(row, origin_row, 5)
# grab the interpolated line
interpolated_line = map_coordinates(m,
np.vstack((y_plot,
x_plot)),
order=1, mode='nearest')
m_max[row][col] = max(interpolated_line)
m_dist[row][col] = np.argmax(interpolated_line)
print(m)
print(m_max)
print(m_dist)
As you can see this is very brute force, and I have managed to broadcast all the code around this part but stuck on this part. here is an illustration of what I am trying to achieve, I will go through the first iteration
1.) the input array
2.) the first loop from 0,0 to origin (3,3)
3.) this will return [10 9 9 8 0] and the max will be 10 and the index will be 0
5.) here is the output for the sample array I used
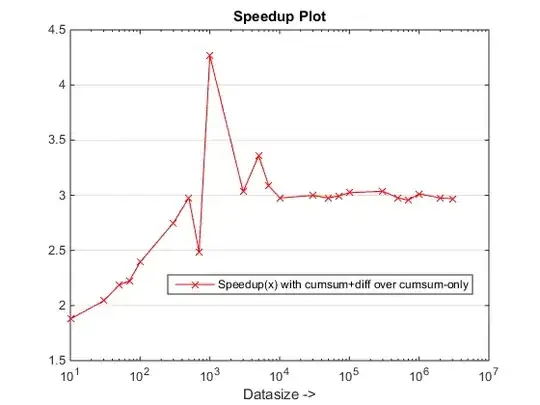
Here is an update of the performance based on the accepted answer.