I have found the DiagrammeR package easiest to do this. The general idea would be something like:
library(glue)
library(DiagrammeR)
excluded <- glue('Full text articles excluded
n = 1000
Reasons for exclusion
Reason 1
Reason 2')
grViz("
digraph cohort_flow_chart
{
node [fontname = Helvetica, fontsize = 12, shape = box, width = 4]
a[label = 'Records identified in original search']
b[label = 'Records identified with update']
c[label = 'Records after duplicates removed']
d[label = 'Records screened']
e[label = 'Records excluded']
f[label = 'Full text articles assessed']
g[label = 'Studies included']
h[label = '@@1']
{ rank = same; a b}
{ rank = same; d, e}
{ rank = same; f, h}
a -> c;
b -> c;
c -> d;
d -> e [ minlen = 3 ];
d -> f;
f -> h [ minlen = 3 ];
f -> g;
}
[1]: excluded
")
Will look like:
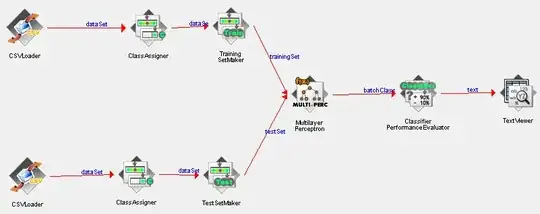
Image with labels and empty nodes
grViz("
digraph cohort_flow_chart
{
node [fontname = Helvetica, fontsize = 12, shape = box, width = 4]
i[label = 'Identification', fillcolor = LightBlue,
style = filled, width = 2]
j[label = 'Screening',fillcolor = LightBlue, style = filled, width = 2]
k[label = 'Eligibility', fillcolor = LightBlue, style = filled,
width = 2]
l[label = 'Included', fillcolor = LightBlue, style = filled, width = 2]
a[label = 'Records identified in original search']
b[label = 'Records identified with update']
c[label = 'Records after duplicates removed']
d[label = 'Records screened']
e[label = 'Records excluded']
f[label = 'Full text articles assessed']
g[label = 'Studies included']
h[label = '@@1']
blank_1[label = '', width = 0.01, height = 0.01]
blank_2[label = '', width = 0.01, height = 0.01]
blank_4[label = '', width = 4, color = White]
{ rank = same; a b i}
{ rank = same; blank_4 c j}
{ rank = same; f k}
{ rank = same; g l}
{ rank = same; blank_1 e}
{ rank = same; blank_2 h}
a -> c;
b -> c;
b -> blank_4 [ dir = none, color = White];
c -> d;
d -> blank_1 [ dir = none ];
blank_1 -> e [ minlen = 3 ];
blank_1 -> f;
f -> blank_2 [ dir = none ];
blank_2 -> h [ minlen = 3 ];
blank_2 -> g;
}
[1]: excluded
")