At the moment I am fitting a Gaussian or Lorentz to my data but both don't fit well enough and I want to switch to Voigt fitting, a convolution of them both.
I retrieved the Voigt function from https://www.originlab.com/doc/Origin-Help/Voigt-FitFunc and wrote it into python.
import numpy as np
import matplotlib.pyplot as plt
def lorentz_voigt(x, A, xc, wL):
return (2*A / np.pi) * (wL / ((4*(x.astype(float) - xc)**2) + wL**2))
def gauss_voigt(x, wG):
return np.sqrt((4*np.log(2)) / np.pi) * ((np.exp(-(((4*np.log(2)) / (wG**2))*(x.astype(float))**2))) / (wG))
def Voigt(x, xc, A, wG, wL):
return np.convolve(lorentz_voigt(x, A, xc, wL), gauss_voigt(x, wG), 'same')
symx = np.linspace(-100, 100, 1001)
asymx = np.linspace(0, 100, 1001)
symy = Voigt(symx, 50, 1, 5, 5)
asymy = Voigt(asymx, 50, 1, 5, 5)
plt.clf()
plt.plot(symx, symy)
plt.plot(asymx, asymy)
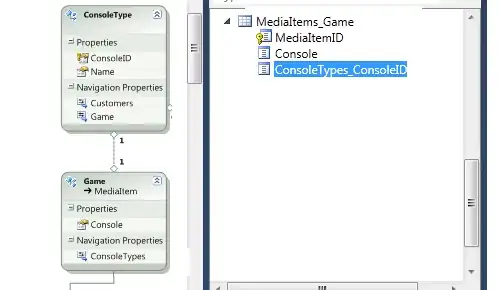
As seen in the plot below the Voigt function with an asymmetric x-axis input, in orange, does not reproduce the correct Voigt profile as seen in blue.
My data is in wavenumbers from 600-4000 cm-1 and I would like to know if I would have to add zeros to my data from -4000 to 600 cm-1 because this is a pure mathematical limitation of convolution or is there a mistake in/solution for my code?