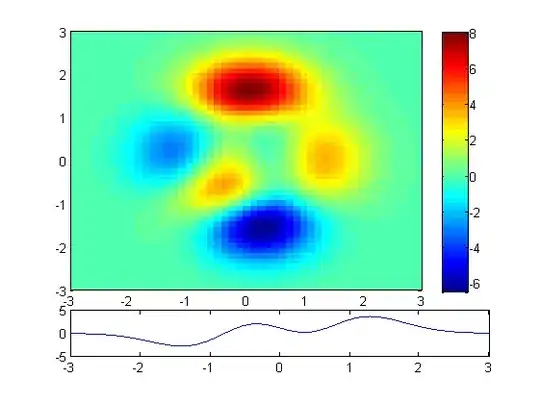
Based on merv's good answer, to answer to myself, I think that closed solution is:
p(yi|α,β,ni,xi)∝ [logit−1(α+βxi)]y * [1 − logit−1(α+βx)n−y]
Thus posterior can be calculated following:
import numpy as np
from scipy import optimize, stats
import matplotlib.pyplot as plt
x = np.array([-0.86, -0.30, -0.05, 0.73])
n = np.array([5, 5, 5, 5])
y = np.array([0, 1, 3, 5])
ngrid = 100
mu_1, mu_2, sd_1, sd_2 = 0, 10, 2**2, 10**2
A = np.linspace(-4, 4, ngrid)
B = np.linspace(-10, 30, ngrid)
mu = np.array([0, 10])
s = np.array([[22, 102]])
Rho = np.array([[1, 0.5], [0.5, 1]])
Sigma = Rho * np.outer(s, s)
prior = stats.multivariate_normal([mu_1, mu_2], Sigma)
def prop_likelihood(input_values):
ilogit_abx = 1 / (np.exp(-(input_values[...,0][...,None]*np.ones(x.shape) + input_values[...,1][...,None] * x)) + 1)
return np.prod(ilogit_abx**y * (1 - ilogit_abx)**(n - y), axis=ilogit_abx.ndim -1)
grid_a , grid_b = np.meshgrid(A,B)
grid = np.empty(grid_a.shape + (2,))
grid[:, :, 0] = grid_a
grid[:, :, 1] = grid_b
posterior_density = prior.pdf(grid)*prop_likelihood(grid)
Which then can be illustrated:
fig, ax = plt.subplots(figsize=(10, 5)
ax.imshow(
posterior_density,
origin='lower',
aspect='auto',
extent=(A[0], A[-1], B[0], B[-1])
)
ax.set_xlim([-4, 4])
ax.set_ylim([-10, 30])
ax.set_xlabel(r'$\alpha$')
ax.set_ylabel(r'$\beta$')
ax.set_title('Posterior heatmap')
ax.grid('off')

Analytical solution:
def opt(params):
a, b = params[0], params[1]
z = np.exp(a + b * x) / (1 + np.exp(a + b * x))
e = - np.sum(y * np.log(z) + (n - y) * np.log(1 - z))
return e
optim_res = optimize.minimize(opt, np.array([0.0, 0.0]))
mu_opt = optim_res['x']
sigma_opt = optim_res['hess_inv']
posterior_optimized = stats.multivariate_normal(mean=mu_opt, cov=sigma_opt)
Which can then be plotted
fig, ax = plt.subplots(figsize=(10, 5)
ax.imshow(
posterior_optimized.pdf(grid),
origin='lower',
aspect='auto',
extent=(A[0], A[-1], B[0], B[-1])
)
ax.set_xlim([-4, 4])
ax.set_ylim([-10, 30])
ax.set_xlabel(r'$\alpha$')
ax.set_ylabel(r'$\beta$')
ax.set_title('Posterior heatmap from analytical solution')
ax.grid('off')

There are some differences. Not sure if analytical optimization function is correct.
Hopefully this helps others.