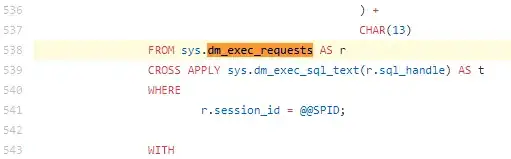
The Variant Schema used by Google Genomics Variant Transform pipelines represents genotypes as nested records in BigQuery - for example:
(from: https://bigquery.cloud.google.com/table/genomics-public-data:1000_genomes.variants?pli=1&tab=preview)
I'm having trouble understanding how to write queries that involve relationships between samples - such as:
select all variants where sampleA.genotype=HET and sampleB.genotype=HET and sampleC.genotype=HOM-ALT
or similar queries where sampleA and sampleB are parents of sampleC and you're looking for variants that follow a particular inheritance pattern.
How are people writing these queries with the nested schema?