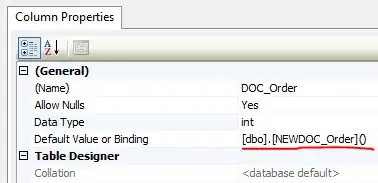
I have done a regression tree with rpart to assess the walking of elderly people based on a few variables. With the use of the plot I would like to use the output for further analysis in another software. However I was wondering whether it would be possible not only derive the walking per group from the leaf nodes, but also to derive the standard deviations from the leaf nodes (in terms of walking)?
#### Decision tree with rpart
modelRT <- rpart(logwalkin~.-walkinmin-walkingtime, data=trainDF,
control=rpart.control(minsplit=25, maxdepth = 8, cp =0.00005))
rpart.plot(modelRT,type=3,digits=3,fallen.leaves=TRUE)