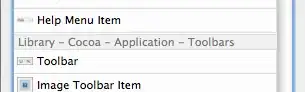
I need to replicate a binomial test from R to SAS but I'm obtaining different results (or maybe I am misinterpreting the SAS results).
In order to explain my problem in an easy way, I will use data from this wikipedia example because it provides the final solution;
Suppose you want to calculate the probability of obtaining 51 or more 6s in a sample of 235 roll of a fair die with 6 faces, so that the true probability of rolling a 6 on each trial is 1/6. The final solution should be 0.02654.
In R, the code to do is the following:
binom.test(51,235,(1/6),alternative = "greater")
and the obtained results are:
Exact binomial test
data: 51 and 235 number of successes = 51, number of trials = 235,
p-value = 0.02654
alternative hypothesis: true probability of success is greater than 0.1666667
95 percent confidence interval:
0.1735253 1.0000000
sample estimates: probability of success
0.2170213
When in SAS the equivalent should be given by:
DATA DICEROLL;
ROLL=51;
FREQQ=235;
PROB=1/6;
RUN;
data _null_;
set diceroll;
call symput("probability",prob);
run;
PROC FREQ DATA=DiceRoll ;
TABLES FREQQ / BINOMIAL (P=&probability.) ALPHA=0.05;
EXACT BINOMIAL ;
WEIGHT ROLL ;
RUN;
But THIS is the results I obtain (in which there is no p-value = 0.02654)
I tried in several ways to reconcile my results(tried all the three alternatives in R, tried to invert ROLL and FREQQ in sas because I wasn't sure) but I still haven't found a solution. Do binom.test and proc freq + BINOMIAL perform at least the same test? Am I misinterpreting the SAS output?
Thank you in advance for you precious help!
============================== UPDATE ============================
I tried both proposed methodology by reeza and BEMR and I feel I am close to the solution! @BEMR: as I wrote and explained bettere in the comment, how should I adapt %r(1,6) if my variable is dichotomic? Your code works with the example of a 6-faced die but in my real case, my success variable assume values between 0 and 1, so I am not sure of what I have to do (I apologize if I did not mention it at the beginning)
@REEZA: Your solution seems to work but I had to remove the /2; I guess your first solution calculates p-values as a two sided test not one sided.
Anyway, the results are fine but there are huge differences between SAS and R when the number of success is 0 or close to 0 (1,2,3). Do you know any workaround for this? Or better, is it safe to assume that the test is unreliable in both cases?
The following pictures are my results with the reeza method, thank you all for your precious cooperation!