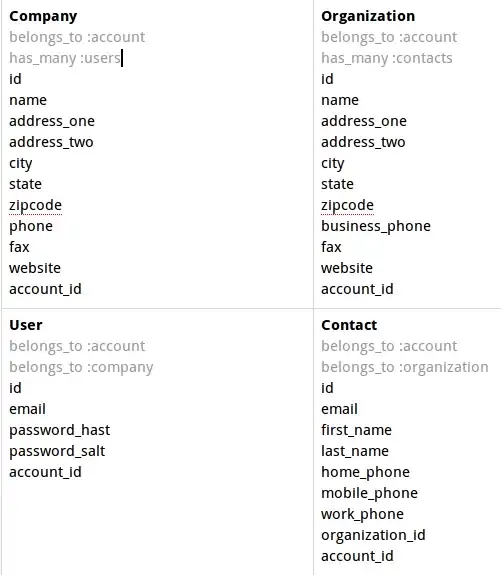
I'm trying to map cis-elements to the promoter regions of the genes of interest we have.
I don't have a database I'm working from, but I have a list from the new-place, sogo website; outputs format into
Factor site, Loc, Str, SITE
So I have the position and strand. Are there any useful R packages for configuring my data into the image shown above, where you can map to the positive and negative strand and can call the SITE as the description for marker in the promoter regions? Ultimately it would be best to configure a legend indicating the different promoter regions by color.