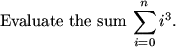I have problems with using pandas for pyplot. On the one hand the scale is wrong, since the value 10 on the y axis shows before 1.
On the other hand I get the error message:
TypeError: unsupported operand type(s) for -: 'str' and 'str'
When using yerr.
import matplotlib.pyplot as plt
import numpy as np
import matplotlib
import pandas as pd
df=pd.read_table('TI_attachment.dat', header=0, sep='\s+')
fig, ax=plt.subplots(figsize=(20,10))
ax.errorbar(x=df.iloc[:, 0:1], y=df.iloc[:, 1:2], yerr=df.iloc[:, 2:3], color='black')
ax.set_xlabel('Simulation Time per window [ns]', size=25)
ax.set_ylabel('Free energy of binding [kcal/mol]', size=25)
ax.tick_params(axis='both', labelsize=25)
plt.tight_layout()
#plt.savefig('PMF.png', format='png')
#plt.show()
This is what TI_attachment.dat looks like:
#Weight of restraints (%), Accumulative work (in kcal/mol), SEM (in kcal/mol)
0.0000 0.00000 0.00000
0.0040 3.23161 0.78401
0.0080 3.76232 0.79356
0.0160 4.50989 0.82542
0.0240 4.86168 0.82490
0.0400 5.48672 0.82894
0.0550 6.02476 0.82931
0.0865 6.73611 0.83116
0.1180 7.20339 0.83305
0.1810 7.69373 0.83432
0.2440 8.16010 0.83487
0.3700 8.87930 0.83952
0.4960 9.25889 0.84035
0.7480 9.83864 0.84071
1.0000 10.28260 0.84107