I have a time series for which I want to intelligently interpolate the missing values. The value at a particular time is influenced by a multi-day trend, as well as its position in the daily cycle.
Here is an example in which the tenth observation is missing from myzoo
start <- as.POSIXct("2010-01-01")
freq <- as.difftime(6, units = "hours")
dayvals <- (1:4)*10
timevals <- c(3, 1, 2, 4)
index <- seq(from = start, by = freq, length.out = 16)
obs <- (rep(dayvals, each = 4) + rep(timevals, times = 4))
myzoo <- zoo(obs, index)
myzoo[10] <- NA
If I had to implement this, I'd use some kind of weighted mean of close times on nearby days, or add a value for the day to a function line fitted to the larger trend, but I hope there already exist some package or functions that apply to this situation?
EDIT: Modified the code slightly to clarify my problem. There are na.* methods that interpolate from nearest neighbors, but in this case they do not recognize that the missing value is at the time that is the lowest value of the day. Maybe the solution is to reshape the data to wide format and then interpolate, but I wouldn't like to completely disregard the contiguous values from the same day. It is worth noting that diff(myzoo, lag = 4) returns a vector of 10's. The solution may lie with some combination of reshape, na.spline, and diff.inv, but I just can't figure it out.
Here are three approaches that don't work:
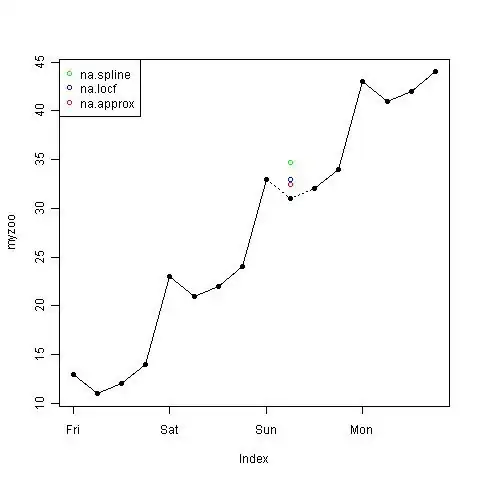
EDIT2. Image produced using the following code.
myzoo <- zoo(obs, index)
myzoo[10] <- NA # knock out the missing point
plot(myzoo, type="o", pch=16) # plot solid line
points(na.approx(myzoo)[10], col = "red")
points(na.locf(myzoo)[10], col = "blue")
points(na.spline(myzoo)[10], col = "green")
myzoo[10] <- 31 # replace the missing point
lines(myzoo, type = "o", lty=3, pch=16) # dashed line over the gap
legend(x = "topleft",
legend = c("na.spline", "na.locf", "na.approx"),
col=c("green","blue","red"), pch = 1)