I have the following dataset. Note that these are fictitious values:
Farm CO2Fert CO2Manure CO2Feed CO2Housing CO2Fuel CO2Other
Best 2.187635996 0.670289261 0.773605214 2.415801361 1.180859203 2.876050311
Worst 4.240789564 0.503797793 1.884548328 5.114791701 3.411847194 5.150085802
User 1.189743632 2.852713125 0.419821994 0.630429463 2.982960729 1.489036959
I have been asked to draw a spider diagram of these values. Having looked at fmsb and ggradar I have for a number of reasons decided to start from scratch instead. So, I have the following code (not currently sure if all the libraries are necessary):
library(dplyr)
library(ggplot2)
library(scales)
library(reshape2)
library(tibble)
library(data.table)
data <- fread("C:/myData/ExampleFootprintData.csv")
dat2test <- melt.data.table(data, c("Farm"), c("CO2Fert", "CO2Manure", "CO2Feed", "CO2Fuel", "CO2Housing", "CO2Other"))
thePlot <- ggplot(data = dat2test, aes(x=variable, y=value, group=Farm, color= Farm)) + ## Define data and colour column
geom_polygon(fill = NA) + ## make the lines meet
coord_polar() ## Make it round
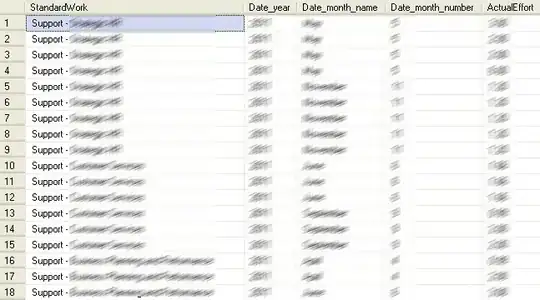
As you can see the labels are not fully onto the plot surface. Looking at the examples around on the internet (e.g. the main text I've taken my code from) this appears not to be addressed anywhere. There is an interesting solution to this here, and I am considering using it, but my eventual labels are likely to be long and descriptive. I would like them to be shown horizontally as they are now with line breaks as appropriate, but so that they fit on the chart.
So far, I have not got very far with this. using theme(axis.title.x = element_text(vjust=-0.5)) or similar does not work, but if I use margins as suggested here, e.g. theme(axis.text.x = element_text(margin = margin(t = -20))) all that changes is that the x axis title moves inwards into the plot area which expands, but the labels are still outside of the plot:
How do I force ggplot to keep my labels inside the physical area of the plot?
dput of the data:
structure(list(Farm = c("Best", "Worst", "User"), CO2Fert = c(2.187635996,
4.240789564, 1.189743632), CO2Manure = c(0.670289261, 0.503797793,
2.852713125), CO2Feed = c(0.773605214, 1.884548328, 0.419821994
), CO2Housing = c(2.415801361, 5.114791701, 0.630429463), CO2Fuel = c(1.180859203,
3.411847194, 2.982960729), CO2Other = c(2.876050311, 5.150085802,
1.489036959), NO3Fert = c(2.19509301, 1.848317101, 1.643695528
), NO3Manure = c(2.452857906, 3.153028268, 1.922113286), NO3Grazing = c(0.037698451,
4.452847769, 2.546101867), NH4Spreading = c(2.880954824, 2.60492997,
3.186211336), NH4Storage = c(1.178815284, 4.893388111, 2.432823901
), NH4Grazing = c(0.509207305, 2.998872111, 4.444466334), NH4Housing = c(2.523406518,
5.255666955, 1.287199958)), .Names = c("Farm", "CO2Fert", "CO2Manure",
"CO2Feed", "CO2Housing", "CO2Fuel", "CO2Other", "NO3Fert", "NO3Manure",
"NO3Grazing", "NH4Spreading", "NH4Storage", "NH4Grazing", "NH4Housing"
), row.names = c(NA, -3L), class = c("data.table", "data.frame"
), .internal.selfref = <pointer: 0x0000000000120788>)