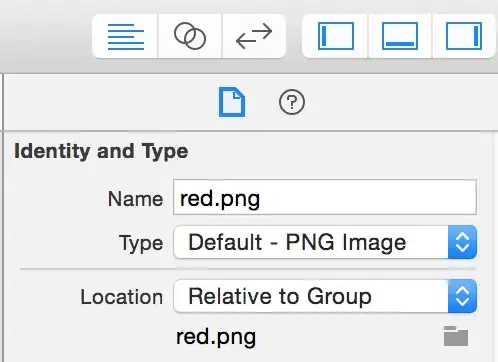
I had the same issue when plotting class decision boundaries (colors) with uncertainty (blending color to white). Between two boundaries I had colors of other classes due to interpolation. These boundaries are wrong. Look at the transition from blue to red:
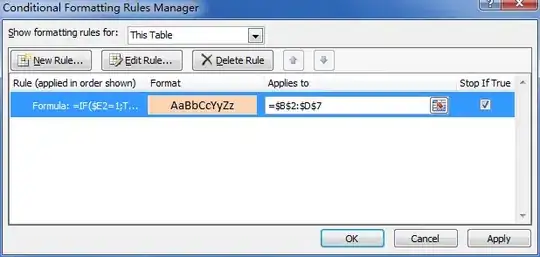
I solved this by plotting only one class at a time. This can be done with masked arrays:
# C contains classes 0, 1, 2 as mesh
# CU contains classes with uncertainty, CU = 2 * C + U, where 0 <= U <= 1
# xx and yy are the mesh variables
for cl in np.unique(C): # iterate over red, green, blue classes
mask = C != cl
xx_ma = np.ma.MaskedArray(xx, mask)
yy_ma = np.ma.MaskedArray(yy, mask)
CU_ma = np.ma.MaskedArray(CU, mask)
cnt = plt.contourf(xx_ma, yy_ma, CU_ma, vmin=0, vmax=5,
cmap=cmap, levels=len(np.unique(CU)))
This is the result:
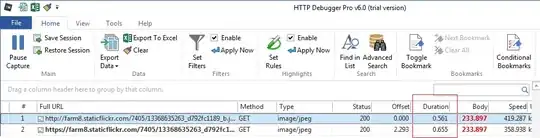
When using pcolormesh, you don't need to plot the classes separately, but the data must be in a different format. Basically you need an xy-mesh for the corners (grid) and another xy-mesh for the centers (values):
(X[i+1, j], Y[i+1, j]) (X[i+1, j+1], Y[i+1, j+1])
+--------+
| C[i,j] |
+--------+
(X[i, j], Y[i, j]) (X[i, j+1], Y[i, j+1]),
The result looks like this (with precise corners now):