I am not professional in Probability & Statisticsin, in order to clearly describe my problem, please be patient of the long introduction.THANKS!
Background of my question
Assume I have several independent random variables, say X and Y, and their distributions are already known, for example: X~N(0,1)$ and $y~N(0,1)$.
Now there is a random function of X and Y, $$g(X,Y)=X^2+Y^2$$, g(X,Y) is a random variable too, make Z=g(X,Y), the distribution of g(X,Y) is unknown, mark it as  . My goal is to get the expectation of g()'s distribution which is E[Z].
. My goal is to get the expectation of g()'s distribution which is E[Z].
Here, the g(X,Y) has a easy form, I can tell g(X,Y) 's distribution is chi2(2). But when the form of g() is more complex, I can't get the distribution of g() directly, more difficult to get the expectation by g(X,Y)'s distribution. --BUT--, I found a web page on wiki which talking about "Law of the unconscious statistician". Then I realized I can get the expection of g()'s distribution by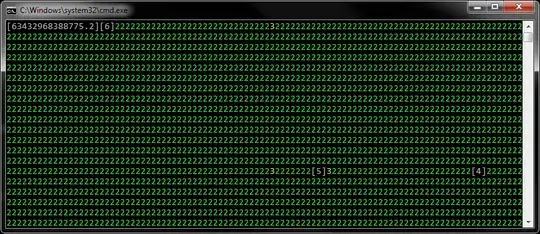 :,
here in this example, the joint distribution:
:,
here in this example, the joint distribution: and
and  .
.
For the equation of E[Z], I think the MC(Monte Carlo) method is a suitable way to deal with it. I rewite the equation based on importance sampling:
h(x,y) is the proposal distribution which has know form. This is a great form to use Metropolis-Hasting Method.
What I want to test is: the expectation calculated by samples from chi2(2) distribution should be equal to the expectation calculated by MC method. I implement all above in python as shown blew:
import numpy as np
import time
import matplotlib.pyplot as plt
import numpy as np
import scipy.stats as st
from scipy.stats import chi2
%matplotlib inline
# np.random.seed(123)
def sampler(mean=[0,0], cov=[[1,0],[0,1]], n=10):
return st.multivariate_normal(mean, cov).rvs(n)
def g(point):
return point[0]**2+point[1]**2
k=2
samples_from_chi2 = chi2.rvs(k,loc=0, scale=1, size=400)
mean_chi2 = np.mean(samples_from_chi2)
iterN = 10000
begin = np.array([2**.5, 2**.5])
samples = np.array(begin)
mulN_mean = np.array([0,0])
mulN_cov = np.array([[1,0],[0,1]])
multiNormal = st.multivariate_normal
p = multiNormal(mulN_mean, mulN_cov)
timeTest_begin = time.clock()
for i in range(iterN):
#print("------------round{a:2d}---------------".format(a=i))
father = begin
pdf_father = p.pdf(father)
son = multiNormal(father, mulN_cov).rvs(1)
pdf_son = p.pdf(son)
# print("father={a:%f}, son={b:%f}".format(a=father, b=son))
'''
in this example, P(father|son) == P(son|father) is true,
so just ignore the conditional probability of Metropolis–Hastings
algorithm
'''
# q_son_father = multiNormal(father, mulN_cov).pdf(son)
# q_father_son = multiNormal(son, mulN_cov).pdf(father)
# print("q(son|father)=", q_son_father)
# print("q(father|son)=", q_father_son)
g_father = g(father)
g_son = g(son)
r_up = g_son * pdf_son
r_down = g_father * pdf_father
r = r_up / r_down
u = np.random.rand()
if r >= u:
samples = np.vstack((samples, son))
else:
samples = np.vstack((samples, father))
begin = samples[-1]
timeTest_end = time.clock()
T = timeTest_end - timeTest_begin
#print("cost time:%d\n", T)
# print("samples=\n", samples)
G_samples = samples[:,0]**2 + samples[:,1]**2
mean_from_sample = np.mean(G_samples)
print("chi2 mean:{a} sample_mean:{b}".format(a=mean_chi2,
b=mean_from_sample))
x = range(iterN+1)
y_max = np.max(G_samples)
y = np.linspace(0, y_max, int(y_max*40))
fig, (ax1, ax2) = plt.subplots(2,1)
ax1.plot(x,G_samples,'-')
ax1.set_title('Trace plot')
ax1.set_xlabel('Number of points')
ax1.set_ylabel('Points location')
ax2.hist(G_samples, int(iterN/100),normed=1)
ax2.set_title('Histogram plot')
ax2.set_xlabel('Points location')
ax2.set_ylabel('Probability')
ax2.plot(y,st.chi2.pdf(2, y), 'r')
fig.subplots_adjust(left=None, bottom=None, right=None, top=None,
wspace=None, hspace=0.8)
The result of the implementation:
chi2 mean:2.0085551240837773 sample_mean:4.069302525056825
Here my question comes
No matter the number of sample points is 400 or 10000, the mean of the samples directly from chi-square distribution is about 2 (I confirm it is right, because if the freedom of chi2 distribution is k, then the expectation is k, and the variance is 2k), but the expectation calculated by MC method is about 4, just as shown above, I want to know why there is a 2 times relationship between two results, if the const's exist is right, how to find it's value when I don't know the random variable function's distribution(in this example, chi-square distribution)
Thanks for reading here, please give me some advice, thank you very much!
Update
the problem has been solved. My assumption has been confirmed: the result by tow ways are equal, and the M-H sampling way is more faster. Here is the new picture of the M-H way(the red line is the pdf plot of $\chi^2(2)$), and mean values of the expectation calculated by tow ways.
chi2 mean:2.0159110138904883 sample_mean:1.9693578501381137