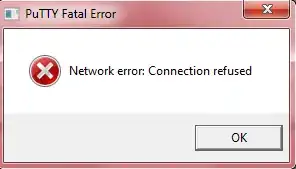
How does one add metadata to a tibble?
I would like a sentence describing each of my variable names such that I could print out the tibble with the associated metadata and if I handed it to someone who hadn't seen the data before, they could make some sense of it.
as_tibble(iris)
# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fctr>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5.0 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
# ... with 140 more rows
# Sepal.length. Measured from sepal attachment to stem
# Sepal.width. Measured at the widest point
# Petal.length. Measured from petal attachment to stem
# Petal.width. Measured at widest point
# Species. Nomenclature based on Integrated Taxonomic Information System (ITIS), January 2018.
thanks!