my question is very simple: i have a database that is looking like this:
My goal is just to eliminate the newline \n at the end of every sequence line, NOT OF THE HEADER, i tried the following code
#!/usr/bin/perl
use strict;
my $db = shift;
my $outfile= "Silva_chomped_for_R_fin.fasta";
my $header;
my $seq;
my $kick = ">";
open(FASTAFILE, $db);
open(OUTFILE,">". $outfile);
while(<FASTAFILE>) {
my $currentline = $_;
chomp $currentline;
if ($currentline =~ m/^$kick/) {
$header = $currentline;
} else {
chomp $currentline;
$seq = $currentline;
}
my $path = $header.$seq."\n";
print(OUTFILE $path);
}
close OUTFILE;
close FASTAFILE;
exit;
But instead of having just the sequence line chomped i obtain the following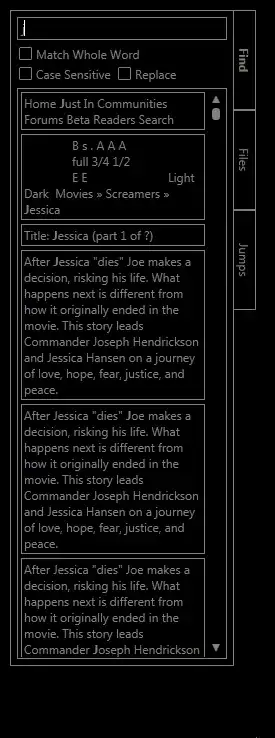
like if chomp didn't work at all.. any idea of what i do wrong? thanks a lot Alfredo