To construct a contractive autoencoder, one uses an ordinary autoencoder with the cost function

To implement this with the MNIST dataset, I defined the cost function using using tensorflow as
def cost(X, X_prime):
grad = tf.gradients(ys=X_prime, xs=X)
cost = tf.reduce_mean(tf.square(X_prime - X)) + tf.reduce_mean(tf.square(grad))
return cost
and used AdamOptimizer for backpropagation. However, the cost doesn't go any lesser than 0.067, which is peculiar. Is my implementation of the cost function incorrect?
Edit:
After reading the documentation ontf.gradients, the above implementation would have computed
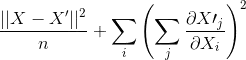 instead. So my question is, how do you do derivatives component wise in tensorflow?
instead. So my question is, how do you do derivatives component wise in tensorflow?

