I am using trackpy to count particles. Working with full images gives satisfying results.
I also have a set of images in which a region is masked (mask and masked image are available here mask+masked image).
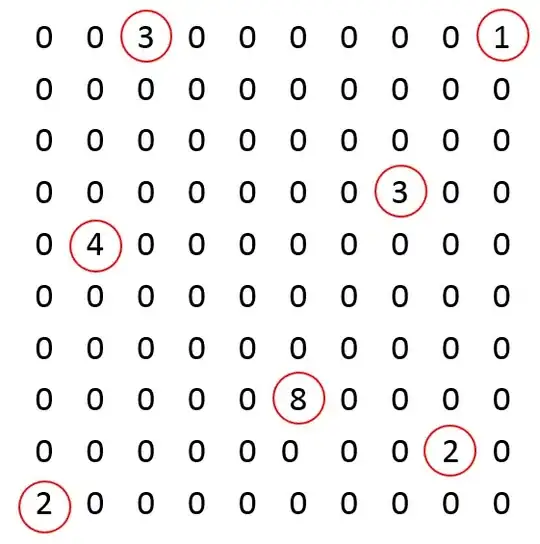
As we can see on the picture bellow, the algorithm detects particles all around the border of the Mask.
import numpy as np
import pandas as pd
from pandas import DataFrame, Series # for convenience
from pylab import*
import pims
import trackpy as tp
frames = pims.ImageSequence('C:/Users/Mich/Desktop/masked_particles.tif', as_grey=True)
originaImage=pims.ImageSequence('C:/Users/Mich/Desktop/masked_particles.tif', as_grey=True)
plt.imshow(frames[0]);
f = tp.locate(frames[0], 3, minmass=30,maxsize=3, separation=3,noise_size=1, threshold=2)
plt.figure(figsize=(40,20)) # make a new figure
tp.annotate(f, originaImage[0],plot_style={'markersize': 1});
What would be the best strategy to prevent this "border-particles" ?
I couldent solve the issue playing with the tp.locate parameters. Maybe there is a way to import the mask and post filter the DataFrame? That would probably be out of my skills.
*****EDIT******
The solution I came up until now consists of modifying the pixel value of the Mask (0) to match the average pixel value of the unmasked region (50 in this example).
The result of particle finding is then less influenced by the border of the Mask even if the resut in not perfect in every case.