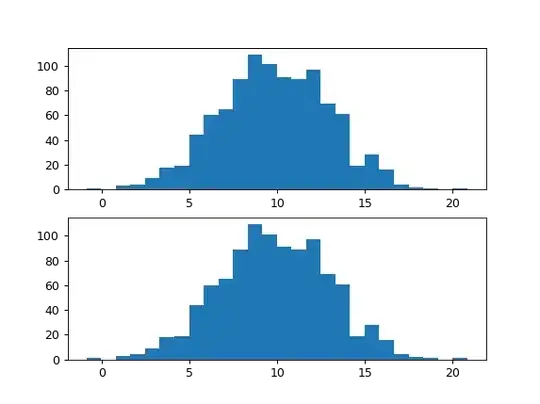
The following lines
a1, b1, _ = plt.hist(df['y'], bins='auto')
a2, b2 = np.histogram(df['y'], bins='auto')
print(a1 == a2)
print(b1 == b2)
equate to all values of a1 being equal to those of a2 and the same for b1 and b2
I then create a plot using pyplot alone (using bins=auto should use the same np.histogram() function):
plt.hist(df['y'], bins='auto')
plt.show()
I then try to achieve the same histogram, but by calling np.histogram() myself, and passing the results into plt.hist(), but I get a blank histogram:
a2, b2 = np.histogram(df['y'], bins='auto')
plt.hist(a2, bins=b2)
plt.show()
From how I understand that plt.hist(df['y'], bins='auto') works, these two plots I am creating should be exactly the same - why isn't my method of using Numpy working?
EDIT
Following on from @MSeifert's answer below, I believe that for
counts, bins = np.histogram(df['y'], bins='auto')
bins is a list of the starting value for each bin, and counts is the corresponding number of values in each of these bins. As shown from my histogram above, this should produce a nearly perfect normal distribution, however, if call print(counts, bins) the result of counts shows that the very first and last bins have quite a substantial count of ~11,000. Why isn't this reflected in the histogram - why is there not two large spikes at either tail?
EDIT 2
It was just a resolution issue and my plot was seemingly too small for the spikes at either end to render correctly. Zooming in allowed them to display.