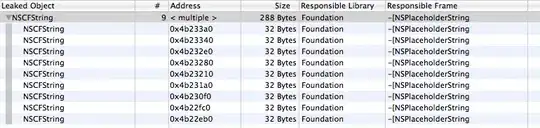
I am trying to take a blank plot, feed it into onRender() from htmlWidgets, and add many lines inside the onRender() function. In the code below, I use a dataset with 100 rows (100 lines), and when I run the application, the 100 lines are drawn inside onRender() in about one second. However, when I change the dataset to have, say, 2000 lines, it will take ten seconds to draw them all.
I am trying to achieve this for datasets on the order of 50,000 to 100,000 lines. This obviously is problematic due to the slowness of the code currently!
The way I am currently achieving the functionality is by:
- Creating a data frame in R called pcpDat. It has 100 rows and 6 columns of numeric data.
- Creating a blank plot in R called p
- Feeding data frame pcpDat and plot p into onRender()
- In onRender(): I have an xArr object that just contains the values 0,1,2,3,4,5. For each row of the data frame, I reconstruct its 6 values into a numeric vector called yArr. Then, for each row of the data, I create a Plotly trace object that contains xArr and yArr to be plotted for the 6 x and 6 y values. This Plotly trace object then creates one orange line for each row of the original data frame.
It may seem silly to have so many lines plotted! My reasoning is I am trying to eventually add functionality so a user can use Plotly to select an area on the plot and view only the lines that intercept that area (the rest of the lines will be deleted). This is why I want the lines to be "interactive".
This all made me ponder a few questions:
- I am not experienced with JavaScript (which is the crux of the onRender() function). I am wondering if it is even possible to expect 50,000 to 100,000 lines to be plotted quickly (within say 5 seconds)?
- If the answer to (1) is that it should be possible, I am seeking advice on how I can "speed up" my code snippet below. Without much JavaScript skills, it is difficult for me to determine what is costing the most time. I could be reconstructing that data inefficiently.
I am eager to hear any advice or opinions on this topic. Thank you!
library(plotly)
library(ggplot2)
library(shiny)
library(htmlwidgets)
library(utils)
ui <- basicPage(
plotlyOutput("plot1")
)
server <- function(input, output) {
set.seed(3)
f = function(){1.3*rnorm(100)}
pcpDat = data.frame(ID = paste0("ID", 1:100), A=f(), B=f(), C=f(), D=f(), E=f(), F=f())
pcpDat$ID = as.character(pcpDat$ID)
plotPCP(pcpDat = pcpDat)
colNms <- colnames(pcpDat[, c(2:(ncol(pcpDat)))])
nVar <- length(colNms)
p <- ggplot(mtcars, aes(x = wt, y = mpg)) + geom_point(alpha=0) + xlim(0,(nVar-1)) +ylim(min(pcpDat[,2:(nVar+1)]),max(pcpDat[,2:(nVar+1)])) + xlab("Sample") + ylab("Count")
gp <- ggplotly(p)
output$plot1 <- renderPlotly({
gp %>% onRender("
function(el, x, data) {
var origPcpDat = data.pcpDat
var pcpDat = data.pcpDat
var Traces = [];
var dLength = pcpDat.length
var vLength = data.nVar
var cNames = data.colNms
xArr = [];
for (b=0; b<vLength; b++){
xArr.push(b)
}
for (a=0; a<dLength; a++){
yArr = [];
for (b=0; b<vLength; b++){
yArr.push(pcpDat[a][cNames[b]]);
}
var pcpLine = {
x: xArr,
y: yArr,
mode: 'lines',
line: {
color: 'orange',
width: 1
},
opacity: 0.9,
}
Traces.push(pcpLine);
}
Plotly.addTraces(el.id, Traces);
}", data = list(pcpDat = pcpDat, nVar = nVar, colNms = colNms))})
}
shinyApp(ui, server)
EDIT: To demonstrate what I am trying to do, I am including 3 images. They show an example where there are 10 rows (lines) in the data. The first image is what the user would see at first (all 10 lines present). Then, the user can use the "Box select" tool and to create a rectangle (gray). Any lines that stay inside the rectangle for all x values it contains remains. In the second image for this example, 5 lines remain. After that, the user can, say, create another rectangle (gray). Again, any lines that stay inside the rectangle for all x values it contains remains. In the third image for this example, only 1 of the lines now remains. These 3 screenshots are from my functioning code. So, I do have a prototype working. However, when I add thousands of lines, it is too slow.