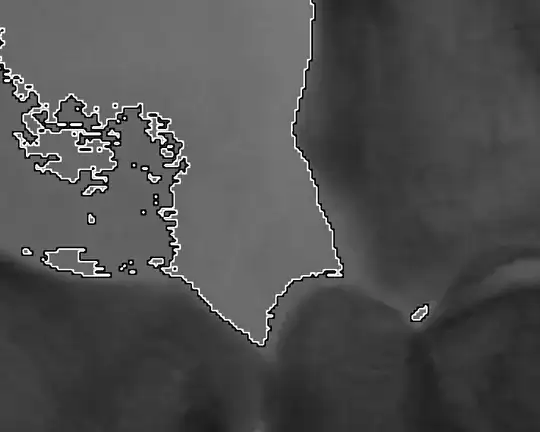
I'm trying to visually inspect, and extract, subsets of large heatmaps. For example, I'd like to roughly extract the row/col indices for clusters like the one I circled below:
Following the advice from here, I hope to achieve this by creating rectangles around subsets of cells by index and repeat until I've highlighted areas close enough to what I want.
Using some simpler data, I tried this:
library(gplots)
set.seed(100)
# Input data 4x5 matrix
nx <- 5
ny <- 4
dat <- matrix(runif(20, 1, 10), nrow=ny, ncol=nx)
# Get hierarchically clustered heatmap matrix
hm <- heatmap.2(dat, main="Test HM", key=T, trace="none")
hmat <- dat[rev(hm$rowInd), hm$colInd]
# Logical matrix with the same dimensions as our data
# indicating which cells I want to subset
selection <- matrix(rep(F,20), nrow=4)
# For example: the third row
selection[3,] <- T
#selection <- dat>7 # Typical subsets like this don't work either
# Function for making selection rectangles around selection cells
makeRects <- function(cells){
coords = expand.grid(1:nx,1:ny)[cells,]
xl=coords[,1]-0.49
yb=coords[,2]-0.49
xr=coords[,1]+0.49
yt=coords[,2]+0.49
rect(xl,yb,xr,yt,border="black",lwd=3)
}
# Re-make heatmap with rectangles based on the selection
# Use the already computed heatmap matrix and don't recluster
heatmap.2(hmat, main="Heatmap - Select 3rd Row", key=T, trace="none",
dendrogram="none", Rowv=F, Colv=F,
add.expr={makeRects(selection)})
This does not work. Here is the result. Instead of the third row being highlighted, we see a strange pattern:
It must have to do with this line:
coords = expand.grid(1:nx,1:ny)[cells,]
# with parameters filled...
coords = expand.grid(1:5,1:4)[selection,]
Can anyone explain what's going on here? I'm not sure why my subset isn't working even though it is similar to the one in the other question.