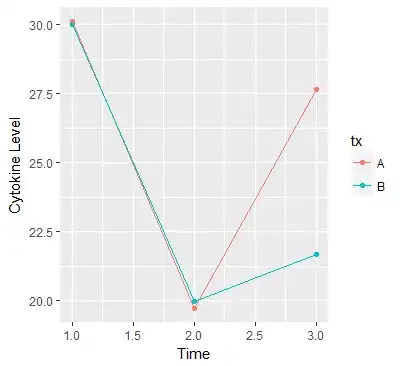
I was trying to plot my longitudinal dataset with ggplot. Here is the example data to illustrate my problem.
library(MASS)
library(ggplot2)
library(dplyr)
# create data
dat.tx.a <- mvrnorm(n=250, mu=c(30, 20, 28),
Sigma=matrix(c(25.0, 17.5, 12.3,
17.5, 25.0, 17.5,
12.3, 17.5, 25.0), nrow=3, byrow=TRUE))
dat.tx.b <- mvrnorm(n=250, mu=c(30, 20, 22),
Sigma=matrix(c(25.0, 17.5, 12.3,
17.5, 25.0, 17.5,
12.3, 17.5, 25.0), nrow=3, byrow=TRUE))
dat <- data.frame(rbind(dat.tx.a, dat.tx.b))
names(dat) = c("measure.1", "measure.2", "measure.3")
dat <- data.frame(subject.id=factor(1:500), tx=rep(c("A", "B"), each=250), dat)
rm(dat.tx.a, dat.tx.b)
dat <- reshape(dat, varying=c("measure.1", "measure.2", "measure.3"),
idvar="subject.id", direction="long")
#ggplot
dat %>%
group_by(tx,time) %>%
summarize(mean=mean(measure), sd=sd(measure)) %>%
ggplot(aes(x=time, y=mean, color=tx))+
geom_point()+
geom_line(aes(group=1))+
labs(x='Time', y='Cytokine Level')
Here is the plot I got. The lines are totally screwed. What was wrong with the plot? thank you.