I know xgboost need first gradient and second gradient, but anybody else has used "mae" as obj function?
3 Answers
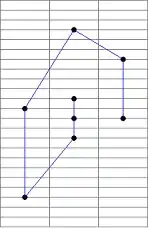
A little bit of theory first, sorry! You asked for the grad and hessian for MAE, however, the MAE is not continuously twice differentiable so trying to calculate the first and second derivatives becomes tricky. Below we can see the "kink" at x=0 which prevents the MAE from being continuously differentiable.
Moreover, the second derivative is zero at all the points where it is well behaved. In XGBoost, the second derivative is used as a denominator in the leaf weights, and when zero, creates serious math-errors.
Given these complexities, our best bet is to try to approximate the MAE using some other, nicely behaved function. Let's take a look.
We can see above that there are several functions that approximate the absolute value. Clearly, for very small values, the Squared Error (MSE) is a fairly good approximation of the MAE. However, I assume that this is not sufficient for your use case.
Huber Loss is a well documented loss function. However, it is not smooth so we cannot guarantee smooth derivatives. We can approximate it using the Psuedo-Huber function. It can be implemented in python XGBoost as follows,
import xgboost as xgb
dtrain = xgb.DMatrix(x_train, label=y_train)
dtest = xgb.DMatrix(x_test, label=y_test)
param = {'max_depth': 5}
num_round = 10
def huber_approx_obj(preds, dtrain):
d = preds - dtrain.get_labels() #remove .get_labels() for sklearn
h = 1 #h is delta in the graphic
scale = 1 + (d / h) ** 2
scale_sqrt = np.sqrt(scale)
grad = d / scale_sqrt
hess = 1 / scale / scale_sqrt
return grad, hess
bst = xgb.train(param, dtrain, num_round, obj=huber_approx_obj)
Other function can be used by replacing the obj=huber_approx_obj.
Fair Loss is not well documented at all but it seems to work rather well. The fair loss function is:
It can be implemented as such,
def fair_obj(preds, dtrain):
"""y = c * abs(x) - c**2 * np.log(abs(x)/c + 1)"""
x = preds - dtrain.get_labels()
c = 1
den = abs(x) + c
grad = c*x / den
hess = c*c / den ** 2
return grad, hess
This code is taken and adapted from the second place solution in the Kaggle Allstate Challenge.
Log-Cosh Loss function.
def log_cosh_obj(preds, dtrain):
x = preds - dtrain.get_labels()
grad = np.tanh(x)
hess = 1 / np.cosh(x)**2
return grad, hess
Finally, you can create your own custom loss functions using the above functions as templates.
Warning: Due to API changes newer versions of XGBoost may require loss functions for the form:
def custom_objective(y_true, y_pred):
...
return grad, hess
- 4,466
- 4
- 29
- 46
-
2Many thanks josh! A quick comment on the log-cosh code; `np.cosh` can overflow for some inputs, using the identity `hess = 1- np.tanh(x)**2` will avoid these issues. – chepyle Jul 23 '18 at 03:10
-
1I think the fair loss function mentioned above is wrong (though the implementation is correct). According to https://www.kaggle.com/c/allstate-claims-severity/discussion/24520, the correct 'Fair Loss' function is given by `y = c * abs(x) - c**2 * np.log(abs(x)/c + 1)`. – kadee Aug 28 '18 at 13:51
-
@kadee Can you share the derivation for your Huber approximations? The LightGBM implementation (https://github.com/Microsoft/LightGBM/blob/5bee6489ac4293328c826e72e7de339206c456da/src/objective/regression_objective.hpp#L278-L293) seems considerably different, with maybe the biggest difference that they set `d` as the negative of how you've defined it. – zkurtz Sep 17 '18 at 02:00
-
@zkurtz - This is the huber approx or psuedo-huber equation. It is an continuously differentiable approximation of the huber loss. This is shown in the link to the huber loss wiki page. I will make this clearer in my answer though. LightGBM might be using some other approx or method. – Little Bobby Tables Sep 18 '18 at 11:42
-
1@josh thanks (and I meant you and not kadee) that makes sense. Finally, could you comment on the answer by @hbar137? Shouldn't you have "d = preds - labels" instead? – zkurtz Sep 18 '18 at 12:42
-
@zkurtz - consider that we are trying to approximate mean _absolute_ error. The absolute error will make all negatives, positive so this doesn't matter in our case. Further, if we are differentiating w.r.t `d` in this case, the fact `d` is negative does not mean that there the first derivative becomes negative. – Little Bobby Tables Sep 18 '18 at 13:34
-
1@josh the line "grad = d / scale_sqrt" is not compatible with "the fact d is negative does not mean that there the first derivative becomes negative". The sign of the gradient is precisely the sign of d, since scale_sqrt is always positive. Empirically, I get vastly different and better results after flipping the sign. – zkurtz Sep 18 '18 at 17:21
-
@zkurtz firstly, let me address the `pred - labels` issue. I believe you are correct in that it should be preds first. This should be true for all of the other functions as well. I will make these changes then review the other queries. – Little Bobby Tables Sep 18 '18 at 19:39
-
@zkurtz - my apologies, my wording was too ambiguous. What I mean is, the fact that `d = (-d_old)` does not mean that `f'(d)` now becomes `-f'(d)`, this is because we are now differentiating w.r.t the new d. – Little Bobby Tables Sep 18 '18 at 20:05
-
4I think that for sklearn you should invert the arguments passed to the objective function... 1st should be labels, 2nd should be predictions... it does not make much difference in this case I suppose, but it can create confusion..also get_labels -> get_label – gabboshow Jan 16 '19 at 13:20
-
@josh: Thanks, it helped me a lot. I have a question which seems a bit off track here. It seems to me that Light GBM still need second derivative in it implement but we can use "MAE" with it as a objective function. Do you have experience on this. Thanks again. – Chau Pham Mar 21 '19 at 03:16
-
1@Catbuilts both xgboost and lightgbm need the 2nd derivative. LighGBM has Fair Loss and Huber Loss out-the-box. If you want more detial I suggest asking a question :) – Little Bobby Tables Mar 21 '19 at 09:25
-
1Upvote since this is very helpful. I implemented this in R (https://github.com/Bixi81/R-ml/blob/master/README.md). I wonder if there is the need to choose a certain type of evaluation error when I use "Huber" or "Fair"? In the moment I evaluate based on MSE. However, since the evaluation error "only" is for updating, the chosen error should not matter too much, right? – Peter Sep 09 '20 at 15:11
-
If you want to optimise for MSE then you can just use MSE/RMSE explicitly as the loss function. In this case, we are approximating MAE with Fair or Huber because we dont have that option. You can use both MSE and MAE to evaluate if you so wish. – Little Bobby Tables Sep 09 '20 at 18:02
-
In my case, I have to set `grad = -d /scale_sqrt` to get normal result for xgb and lgb in sklearn.I test `grad = -d /scale_sqrt` in xgb compared to xgb with `reg:pseudohubererror`, I get the same result in boston datasets. But it seems your definition of grad is the same as [implementation](https://github.com/dmlc/xgboost/blob/33577ef5d33d63c50605097e7895fa6012bce585/src/objective/regression_loss.h#L101) of xgb – Spaceship222 Sep 15 '20 at 09:55
-
1Amazing answer! The latest API seems to have updated the order of arguments, which had me perplexed with my custom objective for a bit: objective4 method signature: `objective(y_true, y_pred) -> grad, hess:` – Kaizzen Oct 10 '22 at 23:53
-
1Thank you @Kaizzen I've added a warning at the bottom of the answer for this :) – Little Bobby Tables Oct 11 '22 at 08:43
For the Huber loss above, I think the gradient is missing a negative sign upfront. Should be as
grad = - d / scale_sqrt
- 39
- 2
-
1Please add comments to the comments section of answers. Also, what makes you think this? I cannot see where the negative comes from. Thanks. – Little Bobby Tables Sep 18 '18 at 13:32
-
If correct, hbar137 deserves upvotes for pointing out something so crucial, slightly more important than your standard comment. – zkurtz Sep 18 '18 at 17:22
-
1@zkurtz it would be if it were correct, but please checkout the [derivative](https://www.math10.com/en/problem-solver/derivative-solver.html?function=c%5E2+*+%28sqrt%28%281+%2B+%28x%2Fc%29%5E2%29%29+-+1%29) of the [psuedo-huber](https://en.wikipedia.org/wiki/Huber_loss#Pseudo-Huber_loss_function). – Little Bobby Tables Sep 18 '18 at 20:16
-
@josh You had the correct formula in the abstract sense. But if you were to put (-x) in place of x, consider that when taking the derivative you would need to apply the chain rule, leading to a minus sign on the derivative. hbar137 was correct at the time they wrote this, but since you fixed your solution (thanks!), this answer is no longer needed. – zkurtz Sep 18 '18 at 20:33
-
I am running the huber/fair metric from above on ~normally distributed Y, but for some reason with alpha <0 (and all the time for fair) the result prediction will equal to zero...
- 3,872
- 3
- 27
- 44