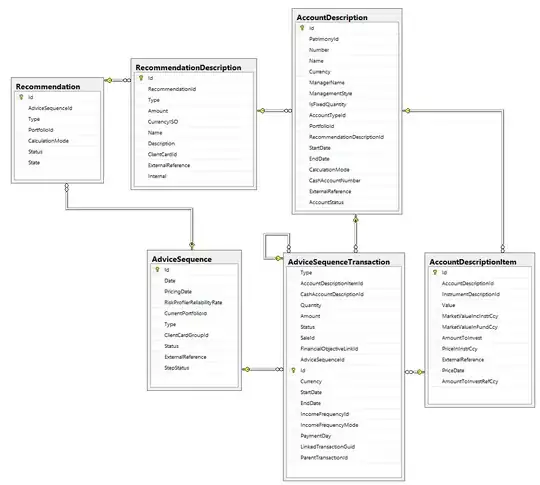
This is the structure of my dataset:
> dput(data)
structure(list(es = c(0.29603085763985, 0.421393627439682, 0.189653473156549,
0.226685054608428, 0.291373762079697, 0.166533544378467, 0.250586529054368,
0.146320008054403, 0.199565119644333, -0.0819047677231083, 0.15963948187092,
-0.154628141843561, 0.201121044198443, 0.0867981239977565, 0.543870310978598,
0.34547921143505, 0.37557241352574, -0.287318919407836, 0.207937483228907,
0.190143660810163, 0.276182673435993, 0.128596803172119, 0.454753165843559,
0.399237234440439, 0.32075358541748, 0.362664873575803, -0.0865925288159671,
0.51290512543514, 0.186308318839249, 0.147936083867325, 0.243792477087184,
0.625169403695832, 0.110317782120045, 0.217836235313289, 0.171468156841181,
0.50548821117127, 0.164418265301427, -0.00246305543239786, 0.325552346507191,
0.381240606108843, 0.19337350462531, 0.0408803528990759, 0.321815078821239,
0.307642815014319, 0.29603085763985, 0.421393627439682, 0.189653473156549,
0.226685054608428, 0.291373762079697, 0.166533544378467, 0.250586529054368,
0.146320008054403, 0.199565119644333, -0.0819047677231083, 0.15963948187092,
-0.154628141843561, 0.201121044198443, 0.0867981239977565,
0.543870310978598,
0.34547921143505, 0.37557241352574, -0.287318919407836, 0.207937483228907,
0.190143660810163, 0.276182673435993, 0.128596803172119, 0.454753165843559,
0.399237234440439, 0.32075358541748, 0.362664873575803, -0.0865925288159671,
0.51290512543514, 0.186308318839249, 0.147936083867325, 0.243792477087184,
0.625169403695832, 0.110317782120045, 0.217836235313289, 0.171468156841181,
0.50548821117127, 0.164418265301427, -0.00246305543239786, 0.325552346507191,
0.381240606108843, 0.19337350462531, 0.0408803528990759, 0.321815078821239,
0.307642815014319, 0.29603085763985, 0.421393627439682, 0.189653473156549,
0.226685054608428, 0.291373762079697, 0.166533544378467, 0.250586529054368,
0.146320008054403, 0.199565119644333, -0.0819047677231083, 0.15963948187092,
-0.154628141843561, 0.201121044198443, 0.0867981239977565, 0.543870310978598,
0.34547921143505, 0.37557241352574, -0.287318919407836, 0.207937483228907,
0.190143660810163, 0.276182673435993, 0.128596803172119, 0.454753165843559,
0.399237234440439, 0.32075358541748, 0.362664873575803,
-0.0865925288159671,
0.51290512543514, 0.186308318839249, 0.147936083867325, 0.243792477087184,
0.625169403695832, 0.110317782120045, 0.217836235313289, 0.171468156841181,
0.50548821117127, 0.164418265301427, -0.00246305543239786, 0.325552346507191,
0.381240606108843, 0.19337350462531, 0.0408803528990759, 0.321815078821239,
0.307642815014319), MAT = c(4.9, 4.9, 15.5, 14.1, 14.1, 14.1,
11.5, 11.5, 11.5, 17, 6.1, 2.7, 2.2, 2.2, 14.1, 14.1, 14.1, 9.5,
9.5, 9.5, 9.5, 9.3, 8.3, 8.266666651, 8.266666651, 4.3, 4.3,
22.3, 14.1, 14.1, 14.1, 8.5, 8.5, 8.5, 8.5, 21.5, 21.5, 3.8,
3.8, 6, 6, 6, 6, 6, 4.9, 4.9, 15.5, 14.1, 14.1, 14.1, 11.5, 11.5,
11.5, 17, 6.1, 2.7, 2.2, 2.2, 14.1, 14.1, 14.1, 9.5, 9.5, 9.5,
9.5, 9.3, 8.3, 8.266666651, 8.266666651, 4.3, 4.3, 22.3, 14.1,
14.1, 14.1, 8.5, 8.5, 8.5, 8.5, 21.5, 21.5, 3.8, 3.8, 6, 6, 6,
6, 6, 4.9, 4.9, 15.5, 14.1, 14.1, 14.1, 11.5, 11.5, 11.5, 17,
6.1, 2.7, 2.2, 2.2, 14.1, 14.1, 14.1, 9.5, 9.5, 9.5, 9.5, 9.3,
8.3, 8.266666651, 8.266666651, 4.3, 4.3, 22.3, 14.1, 14.1, 14.1,
8.5, 8.5, 8.5, 8.5, 21.5, 21.5, 3.8, 3.8, 6, 6, 6, 6, 6), CO2dif = c(162L,
162L, 190L, 165L, 165L, 165L, 200L, 200L, 200L, 150L, 335L, 335L,
335L, 335L, 348L, 348L, 348L, 200L, 200L, 200L, 200L, 220L, 350L,
350L, 350L, 350L, 350L, 350L, 180L, 180L, 180L, 130L, 130L, 130L,
130L, 320L, 320L, 360L, 360L, 345L, 345L, 350L, 348L, 348L, 162L,
162L, 190L, 165L, 165L, 165L, 200L, 200L, 200L, 150L, 335L, 335L,
335L, 335L, 348L, 348L, 348L, 200L, 200L, 200L, 200L, 220L, 350L,
350L, 350L, 350L, 350L, 350L, 180L, 180L, 180L, 130L, 130L, 130L,
130L, 320L, 320L, 360L, 360L, 345L, 345L, 350L, 348L, 348L, 162L,
162L, 190L, 165L, 165L, 165L, 200L, 200L, 200L, 150L, 335L, 335L,
335L, 335L, 348L, 348L, 348L, 200L, 200L, 200L, 200L, 220L, 350L,
350L, 350L, 350L, 350L, 350L, 180L, 180L, 180L, 130L, 130L, 130L,
130L, 320L, 320L, 360L, 360L, 345L, 345L, 350L, 348L, 348L)), row.names = c(NA,
-132L), class = "data.frame", .Names = c("es", "MAT", "CO2dif"
))
I want to create a faceted ggplot in which each facet contains the same points in the background, but each facet has a line with different intercepts among facets. In other words, the slopes are all the same across facets, but the intercepts are different.
First, I replicate the data number_of_facets-times, in this case 3 times, because each facet plots the same points. Each dataset has a different value along the new variable precplot: 500, 1000 or 1500:
data1 <- data; data1$precplot <- 500
data2 <- data; data2$precplot <- 1000
data3 <- data; data3$precplot <- 1500
Join the three datasets
dataplot <- full_join(data1, data2)
dataplot <- full_join(dataplot, data3)
Now plot:
ggplot(dataplot, aes(x = MAT, y = es, color = CO2dif)) +
geom_point(size = 3) + scale_color_gradient(low = "green", high = "red") +
geom_abline(aes(intercept = -0.1846 + 0.0002 * precplot), slope = 0.0211, color = "red", size = 3) +
facet_wrap(~precplot)
Note that the intercept in geom_abline depends on the value of precplot, however, the resulting plot shows the same line with intercept=0 across the three facets.