I am trying to better understand aspects/implications of the observed versus expected information in the context of maximum likelihood estimation. Part of this involves simulating data. If I generate outcome data from the following logistic model:
set.seed(123)
n <- 5000
c1 <- rnorm(n,3,1.5)
c2 <- rnorm(n,5,1.75)
x <- rnorm(n,1+1.25*c1+1.75*c2,1.5)
p<-1/(1+exp(-(-13.5+log(1.5)*x+log(1.25)*c2+log(1.75)*c2)))
y <- rbinom(n,1,p)
dat<-data.frame(c1,c2,x,y)
Then then, if I understood correctly, this code gives me the observed information matrix:
a<-glm(y~x+c1+c2,data=dat,family=binomial(link="logit"))
solve(vcov(a))
But I can't figure out how to obtain the expected information matrix.
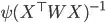 (see
(see