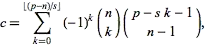I have a function in R that I'm using for creating a map of demographic information.
draw_demographics <- function(map, input, data) {
pal <- colorQuantile("YlGnBu", domain = NULL, n = 7)
#browser()
map %>%
clearShapes() %>%
addPolygons(data = data,
fillColor = ~pal(input$population),
fillOpacity = 0.4,
color = "#BDBDC3",
weight = 1)
}
It's a pure function that takes the map data from Leaflet, the input from the user, and the data from a shapefile to create the map layers. The columns of the shapefile include information like population density, total population, and so on, and I'd like to fill the polygons based on the column name. But where I'm a bit lost is figuring out how to pass selectInput() properly to Leaflet.
Here's a very basic example:
library(shiny)
library(leaflet)
ui <- bootstrapPage(
fluidRow(
column(12, leafletOutput("map"))
),
fluidRow(
column(12, uiOutput("select_population"))
)
)
server <- function(input, output, session) {
output$select_population <- renderUI({
choices <- list("None" = "None",
"All population" = "totalPop",
"Population density" = "totalDens",
"Black population" = "totalAfAm",
"Asian population" = "totalAsian",
"Latino population" = "totalHispanic",
"Native population" = "totalIndian")
selectInput(inputId = "population", label = "Demographics",
choices = choices, selected = "totalDens")
})
output$map <- renderLeaflet({
map <- leaflet() %>%
addProviderTiles(provider = "CartoDB.Positron",
providerTileOptions(detectRetina = FALSE,
reuseTiles = TRUE,
minZoom = 4,
maxZoom = 8)) %>%
setView(lat = 43.25, lng = -94.30, zoom = 6)
map %>% draw_demographics(input, counties[["1890"]])
})
}
## Helper functions
# draw_demographics draws the choropleth
draw_demographics <- function(map, input, data) {
pal <- colorQuantile("YlGnBu", domain = NULL, n = 7)
#browser()
map %>%
clearShapes() %>%
addPolygons(data = data,
fillColor = ~pal(input$population),
fillOpacity = 0.4,
color = "#BDBDC3",
weight = 1)
}
shinyApp(ui, server)
Where I'm a bit lost is how to pass the vector values from the column totalDens from the user's input of totalDens from the dropdown (or, pass whichever column of data they choose to map) to Leaflet. In other words, if a user selects totalPop instead, how can I tell Leaflet to reapply the color palette to this new set of data and re-render the polygons? I attempted using a reactive to get the results of input$population, but to no avail.
Any suggestions, or ways I could troubleshoot? Thanks!