I'm trying to create a time series contourf plot of some velocity/depth data I collected out at sea. The raw data is in a .mat format and I was able to read this into a pandas data frame and produce something really close to what I need, except for the x-axis.
This code produces this plot:
fig = plt.figure()
ax = fig.add_subplot(111)
time, depth = np.meshgrid(a2,b2)
cont = ax.contourf(time, depth, c2, 50, cmap=plt.cm.get_cmap('viridis'),vmin=-1, vmax=1)
ax.set_ylabel("Depth (m)",size=35)
ax.set_xlabel("Time (date YYY/MM/DD)", size=35)
ax.tick_params(labelsize=35)
plt.gca().invert_yaxis()
plt.title("ADCP North-South Velocity", size = 50)
cbar = fig.colorbar(cont)
cbar.set_label(label='Velocity (m/s)', size = 35)
cbar.ax.tick_params(labelsize=35)
mpl.rcParams['figure.figsize'] = 70,35
plt.savefig('adcpV.png')
plt.show()
1st plot
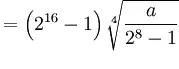
Because it is a matlab file, the time values given are a reference number (i.e. 736456 (days since some reference date) is May 6th, 2016), which I was able to convert into python datetime objects with this:
def matlab2datetime(matlab_datenum):
day = dt.datetime.fromordinal(int(matlab_datenum)) - dt.timedelta(days = 366)
return day
axisdate = []
for elem in a2:
axisdate.append(matlab2datetime(elem))
If I add this line to the first block of code, it produces this plot:
ax.set_xticklabels(axisdate)
2nd plot
In the first plot, the range of the x-axis is about a year, but when I change them into the converted dates, the same date shows up for each tick mark. The reference numbers were all correctly converted to the correct date, but there are some 64,000 data points with multiple data for the same date. So the first few thousand elements in the axisdate list are 2016-05-06, and so on up until 2017-04-19.
First question: How do I get the right date labels for the x-axis?
Second question: How do I remove the hours, minutes, and seconds (00:00:00) from each of the date time objects so that it only shows the day (2016-05-06)?
Here is some of the a2, b2, and c2 data, respectively:
Float64Index([736456.208333, 736456.213542, 736456.21875, 736456.223958,
736456.229167, 736456.234375, 736456.239583, 736456.244792,
736456.25, 736456.255208,
...
736804.703125, 736804.708333, 736804.713541, 736804.71875,
736804.723958, 736804.729166, 736804.734375, 736804.739583,
736804.744791, 736804.75],
dtype='float64', length=64142)
Int64Index([ 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36,
38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66, 68, 70,
72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96],
dtype='int64')
[[ 0.004 0.009 -0.012 ..., -0.072 -0.09 -0.098]
[-0.022 0.013 -0.067 ..., -0.319 -0.293 -0.253]
[-0.053 0.005 -0.017 ..., -0.335 -0.328 -0.343]
...,
[-0.065 0.059 0.05 ..., -0.036 0.018 0.009]
[-0.041 -0.03 -0.035 ..., -0.027 -0.07 -0.046]
[ 0.215 -0.287 0.033 ..., -0.049 0.002 -0.01 ]]